Abstract
The SARS-CoV-2 Omicron variant (B.1.1.529 lineage) escapes antibodies that neutralize the ancestral virus. We tested human serum panels from participants with differing infection and vaccination status using a multiplex surrogate virus neutralization assay targeting 20 sarbecoviruses. We found that bat and pangolin sarbecoviruses showed significantly less neutralization escape than the Omicron variant. We propose that SARS-CoV-2 variants have emerged under immune selection pressure and are evolving differently from animal sarbecoviruses.
Similar content being viewed by others
Main
The coronavirus disease 2019 (COVID-19) pandemic started in December 2019 and has caused 591 million cases and claimed 6.4 million lives as of 19 August 2022. SARS-CoV-2 (ref. 1), which causes COVID-19, is a member of the subgenus Sarbecovirus, as is SARS-CoV-1, which caused the SARS epidemic 19 years ago2. Bats are reservoirs for SARS-related coronaviruses3,4. Multiple sarbecoviruses have been detected in bats5,6,7,8 and, more recently, in pangolins9. SARS-CoV-2 variants of concern (VOC) have emerged since late 2020, probably in response to immune responses in the human population10. So far, five major VOCs have been recognized by the WHO (Alpha, Beta, Gamma, Delta and Omicron). SARS-CoV-2 VOCs have either developed resistance/escape to neutralizing antibodies11,12,13,14,15,16 or acquired mutations that have increased transmission or pathogenicity17,18. SARS-CoV-2 Omicron, which contains 32 amino acid mutations in the spike, was first detected in South Africa and Botswana and has rapidly spread to many countries19,20,21,22.
We applied our SARS-CoV-2 surrogate virus neutralization test (sVNT) platform23,24 which can detect total immunodominant neutralizing antibodies targeting the viral spike (S), and differentiated antibody responses to several human and animal sarbecoviruses in a 20-plex assay that included 15 receptor binding domains (RBDs) of the SARS-CoV-2 clade (clade-1b) viruses and 5 RBDs of the SARS-CoV-1 clade (clade-1a) viruses. Clade-1b viruses included the ancestral SARS-CoV-2 virus (Wuhan-hu-1), variants of concern or interest (Alpha, Delta, Beta, Gamma, Delta plus, Lambda, Mu, Omicron BA.1 and Omicron BA.2) and animal sarbecoviruses (BANAL-52, BANAL-236, GD-1, RaTG13 and GX-P5L). For clade-1a, we included human SARS-CoV-1 and bat sarbecoviruses (Rs2018B, LYRa11, RsSHC014 and WIV1). The phylogenetic relatedness and the amino acid sequence differences of these RBDs and spikes are shown in Extended Data Fig. 1a–d.
All RBDs bind to human ACE2 in a dose-dependent manner in the multiplex Luminex system (Extended Data Fig. 2a). We first tested the 20-plex sVNT using the WHO international standard 20/136 (ref. 25) and found that the neutralization titres were reduced from ancestral SARS-CoV-2 in the following order for the human SARS-CoV-2 variants (from least to most): Alpha, Delta, Delta plus, Lambda, Gamma, Beta, Mu, Omicron BA.2 and Omicron BA.1 (Extended Data Fig. 2b). The calibration of WHO international standards using SARS-CoV-2 ancestral multiplex sVNT was modelled (Extended Data Fig. 2c). From a well-defined panel of 120 sera with varying levels of neutralizing antibodies (NAbs), the data showed a good correlation between sVNT and pVNT (pseudovirus-based VNT) with R2 of 0.83 and 0.73 for the ancestral and Omicron BA.1 virus, respectively, and with good correlation with Omicron BA.1 plaque reduction neutralization test (PRNT) with R2 of 0.79 (Extended Data Fig. 2d–f).
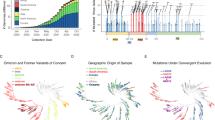
With our 20-plex sVNT platform, we observed significantly more neutralization escape by Omicron BA.1 and BA.2 than by any of the other variants, and this was consistent for all 20 serum panels tested in this study (Fig. 1a) (extent of neutralization in decreasing order: Ancestral > Alpha > Delta > Delta plus > Lambda > Gamma > Beta > Mu > Omicron BA.2 > to lowest Omicron BA.1). Vaccinated individuals who received a third dose of BNT162b2, mRNA-1273 or AZD1222, but not inactivated vaccines, increased overall NAb titre to all viruses. Even in these individuals, a significant reduction in NAb level to Omicron was evident in all serum panels, including those with hybrid immunity (Fig. 1a). Even in those with Omicron-breakthrough infections, the NAb titre to Omicron remained lower than for other SARS-CoV-2 variants (Fig. 1a).
a, A heat map of sVNT GMT50 of 20 serum panels derived from convalescent, vaccinated, boosted and hybrid individuals for immunity against 20 sarbecoviruses using multiplex sVNT. b, Illustration of the clade-1b (SARS-CoV-2, RaTG13 and GX-P5L) and clade-2 (RmYN02 and SL-ZC45) RBDs used in rabbit immunization. The receptor binding motif is highlighted in blue. The percentage of amino acid sequence identity to SARS-CoV-2 RBD is indicated. c,d, The NAb level of the hyperimmune rabbit sera to different sarbecoviruses were determined by 20-plex sVNT (c) and pVNT (d) of different sarbecovirus spike proteins indicated above the panels.
Next, we tested a panel of well-defined hyperimmune rabbit sera raised against different recombinant RBD proteins of clade-1b and clade-2 sarbecoviruses, including human SARS-CoV-2, bat CoV RaTG13, pangolin CoV GX-P5L (clade-1b) and two non-ACE2-binding RBDs from bat CoV RmYN02 and bat CoV SL-ZC45 (clade-2)5. Both RmYN02 and SL-ZC45 RBD have major deletions in the receptor binding motif and shared 62.8% and 65.6% amino acid sequence identity to SARS-CoV-2, respectively (Fig. 1b). Consistent with our data from human sera, Omicron BA.1 and BA.2 are the only SARS-CoV-2 viruses that escape NAbs from all rabbit sera in the panel. Sera raised against RaTG13 and GX-P5L RBDs had 6.4-fold and 3.8-fold NAb titre reduction, respectively, to SARS-CoV-2, but Omicron BA.1/BA.2 showed an almost complete NAb escape with a 58-fold/38-fold and 18-fold/16-fold reduction, respectively, against the RaTG13 and GX-P5L hyperimmune sera (Fig. 1c). This was confirmed by pVNT with a 135-fold reduction of neutralizing activity to Omicron BA.1 in GX-P5L hyperimmune sera (Fig. 1d). The hyperimmune sera raised against the non-ACE2-binding RBDs of RmYN02 and SL-ZC45 had the lowest cross-NAbs to SARS-CoV-2 (Fig. 1c,d). As a negative control, no NAb was detected in the rabbit hyperimmune sera raised against a non-sarbecovirus bat CoV HKU1 RBD (Fig. 1c,d).
When comparing NAb escape between human VOCs and animal sarbecoviruses, we observed that Omicron had greater neutralization escape than was observed in animal sarbecovirus RaTG13 and GX-P5L using multiplex sVNT, although there were only 15–16 amino acid mutations in Omicron RBDs compared with 22 and 30 mutations in RaTG13 and GX-P5L, respectively (Extended Data Figs. 1a,b and 2a,b). In addition to Omicron, we further observed that VOCs Beta and Mu had higher NAb escaping ability than their most genetically related animal sarbecoviruses bat BANAL-52 and pangolin GD-1 (Fig. 2a,b), even though these animal viruses are phylogenetically more distant from SARS-CoV-2 (Extended Data Fig. 1a,c). Using pVNT, we further confirmed that the SARS-CoV-2 Beta variant is more potent in NAb evasion than animal sarbecoviruses BANAL-52 and GD-1 despite the animal sarbecoviruses containing 20 and 127 mutations (Extended Data Figs. 1d and 3a and Fig. 2c,d), respectively, in the spike protein. SARS-CoV-2 Omicron BA.1 and BA.2 have comparable pVNT 50% neutralization titre (NT50) with GX-P5L (Fig. 2c).
a, Effect of RBD mutations examined by multiplex sVNT is shown by neutralization titres (n = 75) for SARS-CoV-2 ancestral, Delta, Beta, Mu, Omicron BA.1, Omicron BA.2, Bat CoV BANAL-52, RaTG13, Pangolin CoV GD-1, GX-P5L and SARS-CoV-1. b, The effect of the number of RBD mutations on NAb escape (n = 89). The effect of all mutations in the full spike protein was measured using pVNT and is shown in c. c, Neutralization titres (n = 75) for SARS-CoV-2 ancestral, Delta, Beta, Omicron BA.1, Omicron BA.2, Bat CoV BANAL-52, Pangolin CoV GD-1, GX-P5L and SARS-CoV-1. d, The effect of the number of spike mutations on NAb escape (n = 69). Statistical analysis in a and c was performed using two-sided Wilcoxon rank sum test in GraphPad Prism 8 (in a: Beta versus GD-1 P < 0.0001, Beta versus BANAL-52 P < 0.0001, Mu versus GD-1 P < 0.0001, Mu versus BANAL-52 P < 0.0001, Omicron BA.1 versus RaTG13 P < 0.0001, Omicron BA.1 versus GX-P5L P < 0.0001, Omicron BA.1 versus Omicron BA.2 P < 0.0001, Omicron BA.2 versus RaTG13 P < 0.0001, and Omicron BA.2 versus GX-P5L P < 0.0001; in c: Beta versus GD-1 P < 0.0001, Beta versus BANAL-52 P < 0.0001, Omicron BA.1 versus GX-P5L P = 0.0035, Omicron BA.1 versus GD-1 P < 0.0001, Omicron BA.1 versus Omicron BA.2 P = 0.0063). Each dot denotes the NT50 value of a sample, while the box shows the interquartile range with median at the centre, and the whiskers represent the maximum and minimum. Linear regression analysis in b and d was performed using GraphPad Prism 8. Error bars in b and d indicate standard deviation of the geometric mean.
Furthermore, SARS-CoV-1 survivors who received two doses of BNT162b2 vaccine displayed broad NAbs against all known sarbecoviruses before the emergence of Omicron. However, sera from this panel also had some degree of NAb escape to Omicron (Fig. 1a). We showed that amino acid mutation G339D, S371L/F, S373P, S375F, D406N and R408S located at the conserved regions are unique to Omicron but are not present in animal sarbecoviruses or SARS-CoV-1 (Extended Data Fig. 3b). Interestingly, S371, S373, S375, D406 and R408 are located in the pan-sarbecoviruses neutralizing epitopes of class VI antibodies26,27. In our mutagenesis data, we observed that mutations at the receptor binding motif (amino acids 417–508) had little-to-no impact on escape from NAbs derived from BNT162b2-vaccinated SARS-CoV-1 survivors, but sera from this panel showed 2.9-fold reduction in geometric mean neutralizing titer 50% (GMT50) against ancestral RBD in RBD with S371L, S373P and S375F mutations (Extended Data Fig. 3c).
Finally, data from antigenic cartography confirmed that Omicron VOCs are antigenically more distinct from the ancestral SARS-CoV-2 than animal sarbecoviruses (Extended Data Fig. 4a). Consistent with our previous findings, we found that the overall antigenic breadth of BNT162b2-vaccinated SARS-CoV-1 survivors is significantly broader than that of BNT162b2-vaccinated infection-naïve individuals (Extended Data Fig. 4b). More recently, Omicron BA.5, which carries three additional mutations compared with BA.2 in the RBD (L452R, F486V and R493Q), emerged and is overtaking Omicron BA.2 in South Africa and the United States. Using the same serum panels, we demonstrated even more potent NAb escape of mRNA vaccine-induced neutralizing antibodies by Omicron subvariants BA.2.11 and BA.5 with the additional L452R mutation and L452R/F486V/R493Q mutations, respectively (Extended Data Fig. 5).
Combining our sVNT detection platform with a collection of 20 different human serum panels, we have shown that the degree of NAb escape in VOCs is greater than that of distantly related animal sarbecoviruses. We propose that the SARS-CoV-2 Omicron variant emerged under immune selection imposed during 2 years of virus transmission in humans. On the other hand, the sarbecoviruses in animals, mainly in bats, seem to be involving at a slower rate probably due to two possibilities. First, ACE2 may not be the only or main entry receptor in bats. Second, antibody-mediated immune responses are not as strong in bats as in humans28.
Methods
Human serum panels
Naïve sera (n = 20) were used as a control. The convalescent panel consists of sera derived from early pandemic (n = 20), Beta-infected (n = 20), Delta-infected (n = 15) and SARS-infected (n = 14) individuals. The vaccinated panel contains sera from individuals who received two doses of COVID-19 vaccine (BNT162b2 (n = 20), mRNA-1273 (n = 20), CoronaVac (n = 20), BBIBP-CorV (n = 17) or AZD1222 (n = 20)), at 14 d post second dose. The boosted panel contains sera from individuals who received a booster after two doses of COVID-19 vaccines, the booster comprising homologous booster: BNT162b2 ×3 (n = 20), mRNA-1273 ×3 (n = 9), CoronaVac ×3 (n = 20), or heterologous booster: BNT162b2 ×2/mRNA-1273 (n = 20), CoronaVac ×2/BNT162b2 (n = 12), CoronaVac ×2/AZD1222 (n = 20) and CoronaVac ×2/BBIBP-CorV (n = 7). The hybrid panel consists of sera from individuals with Delta-breakthrough infections (n = 20), Omicron-breakthrough infections (n = 49), individuals with previous exposure to SARS-CoV-2 with two doses of BNT162b2 (n = 11) and individuals with previous exposure to SARS-CoV-2 with one dose of BNT162b2 (n = 20). Ethics statements, Institutional Review Board approvals and references to previous studies are listed in Supplementary Data Table 1.
Rabbit hyperimmune sera raised against recombinant RBD proteins
Rabbit anti-SARS-CoV-2, GX-P5L, RaTG13, RmYN02, SL-ZC45 and HKU1 RBD sera were all custom-produced by Genscript. Rabbit anti-SARS-CoV-1 sera were described in previous studies29.
Cell line
Lung carcinoma epithelial cells (A549, ATCC CRM CCL-185) were grown and maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS). A549-ACE2 cells were produced by transduction of third-generation lentiviruses carrying human ACE2 gene under EF1-alpha promoter in pFUGW vector. A549-ACE2 cells were maintained in RPMI-1640 supplemented with 10% FBS and 15 µg ml−1 blasticidin.
Enzymatic biotinylation of recombinant RBD proteins
Biotinylated RBD proteins from ancestral SARS-CoV-2, SARS-CoV-2 Alpha, Delta, Beta, Gamma, Bat CoV RaTG13, Pangolin CoV GX-P5L and SARS-CoV-1 were custom-made by Genscript. Biotinylated SARS-CoV-2 Omicron BA.1 RBD was purchased from Acrobiosystems. Biotinylated RBDs from SARS-CoV-2 Delta plus, Mu, Lambda and Omicron BA.2, bat CoVs BANAL-52, BANAL-236, WIV1, Rs2018B, LYRa11 and RsSHC014, and pangolin CoV GD-1 were produced in-house. Briefly, the RBD coding sequences were cloned into pcDNA3.1 vector with SARS-CoV-2 signal peptide (amino acid 1–14) at the N terminus and 10x his-tag, followed by AviTag at the C terminus. After transfection of expression plasmid into HEK293T cells using Fugene6 in Opti-MEM media, expressed proteins were collected at day 3 or day 6 post transfection. RBD proteins were purified using Ni Sepharose (GE Healthcare) and desalted using Amicon Ultra-4, 10 KDa cutoff (Merck). Enzymatic biotinylation of AviTag was performed using BirA protein-biotin ligase kit (Avidity) according to the manufacturer’s instructions. Excessive biotin was removed by Amicon Ultra-4, 10 MW (Merck). Protein concentration was determined by Nanodrop (Thermo Scientific).
Sarbecovirus RBDs for the 20-plex sVNT assay system
The RBDs included in this study are as follows. (1) Clade-1b sarbecoviruses: SARS-CoV-2 ancestral, SARS-CoV-2 VOCs (Alpha, Beta, Gamma, Delta, Omicron BA.1 and Omicron BA.2), SARS-CoV-2 variants of interest (Delta plus, Lambda, Mu), animal sarbecoviruses (bat CoV BANAL-52, pangolin CoV GD-1, bat CoV BANAL-236, bat CoV RaTG135, pangolin CoV GX-P5L); (2) Clade-1a sarbecoviruses: SARS-CoV-1 and bat CoVs WIV1, Rs2018B, LYRa11 and RsSHC014.
Multiplex sVNT
Multiplex sVNTs were established as previously described24. Briefly, AviTag-biotinylated RBD proteins were coated on MagPlex-Avidin microspheres (Luminex) at 5 µg million−1 beads. RBD-coated beads (600 per antigen) were pre-incubated with testing serum at final dilutions of 1:20, 1:80, 1:320 and 1:1,280 for 15 min at 37 °C with agitation, followed by addition of 50 µl R-phycoerythrin-conjugated human ACE2 (2 mg ml−1; Genscript) and incubation for an additional 15 min at 37 °C with agitation. After two washes with 1% BSA in PBS, the final readings were acquired using the MAGPIX system (Luminex) following the manufacturer’s instruction.
pVNT
SARS-CoV-2 Wuhan-hu-1 (ancestral), Delta, Beta, Omicron BA.1, Omicron BA.2, Omicron BA.2.11 (L452R), Omicron BA.2 F486V, Omicron BA.5, GD-1, BANAL-52, GX-P5L and SARS-CoV-1 full-length spike-pseudotyped viruses were produced and packaged as previously described. Briefly, 5 million HEK293T cells were transfected with 20 µg of pCAGGS spike plasmid using FuGene6 (Promega). At 24 h post transfection, cells were incubated with VSV∆G luc seed virus (at a multiplicity of infection of 5) for 2 h. Following two PBS washes, infected cells were replenished with complete growth media supplemented with 1:5,000 diluted anti-VSV-G mAb (Clone 8GF11, Kerafast). At 24 h post infection, pseudoviruses were collected by centrifugation at 2,000 × g for 5 min. For pVNT assay, 3 × 106 relative light units of pseudoviruses were pre-incubated with 4-fold serial-diluted test serum in a final volume of 50 µl for 1 h at 37 °C, followed by infection of ACE2-stably-expressing A549 cells. At 20–24 h post infection, an equal volume of ONE-Glo luciferase substrate (Promega) was added and the luminescence signal was measured using the Cytation 5 microplate reader (BioTek) with Gen5 software version 3.10.
PRNT
SARS-CoV-2 Omicron BA.1 clinical isolates, previously isolated using Vero TMPRSS2 cells30 were used in this study. Briefly, 50 plaque-forming units of the Omicron BA.1 were pre-incubated with 4-fold serial-diluted serum for 1 h at 37 °C after previous inoculation into monolayer A549-ACE2 cells. After 1 h incubation, the inoculum was removed and the cells were overlaid with plaque medium containing DMEM with 2% FBS, 0.2% carboxymethylcellulose and 0.8% avicel. The cells were fixed with 10% buffered formalin and stained with 0.2% crystal violet at 72 h post infection.
Phylogenetic analysis
ACE2-binding sarbecovirus spike or RBD sequences were either directly retrieved from NCBI or translated from nucleotide sequences retrieved from GISAID. Further analysis was performed in Geneious Prime (version 2022.0.2). Spike or RBD protein sequences were aligned with MAFFT to construct the phylogenetic tree by the maximum-likelihood method with the blosum62 model using 1,000 bootstrap replicates in the PHYML 3.0 software.
Antigenic cartography
An antigenic map was generated using the R package ‘Racmacs’ (version 1.1.18) in R (version 4.1.2) on the basis of the matrix of NT50 of sera on the 20 sarbecoviruses on multiplex sVNT. The number of optimizations was set to 1,000.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 software. Differences between groups were analysed using two-sided Wilcoxon rank sum test. Correlations between sVNT and pVNT or PRNT were analysed using Pearson correlation coefficients.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data generated in this study are presented in the article and its Supplementary Information. Data and code on the antigenic cartography are available at https://github.com/Lelouchzhu/OmicronDifferentialEscape. Biological materials including cell lines and plasmids are available on reasonable request from the corresponding authors. Source data are provided with this paper.
References
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Drosten, C. et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 (2003).
Li, W. et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679 (2005).
Lau, S. K. et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl Acad. Sci. USA 102, 14040–14045 (2005).
Wacharapluesadee, S. et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 12, 972 (2021).
Delaune, D. et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat. Commun. 12, 6563 (2021).
Ge, X. Y. et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503, 535–538 (2013).
Hu, B. et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 13, e1006698 (2017).
Lam, T. T. et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583, 282–285 (2020).
Harvey, W. T. et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424 (2021).
Wang, P. et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 29, 747–751 e744 (2021).
Wang, P. et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021).
Chen, R. E. et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 27, 717–726 (2021).
McCallum, M. et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 373, 648–654 (2021).
McCallum, M. et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 374, 1621–1626 (2021).
Planas, D. et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596, 276–280 (2021).
Mlcochova, P. et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599, 114–119 (2021).
Saito, A. et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature https://doi.org/10.1038/s41586-021-04266-9 (2021).
Cao, Y. et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature https://doi.org/10.1038/s41586-021-04385-3 (2021).
Schmidt, F. et al. Plasma neutralization properties of the SARS-CoV-2 Omicron variant. N. Engl. J. Med. 386, 99–601 (2022).
Collie, S., Champion, J., Moultrie, H., Bekker, L. G. & Gray, G. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa. N. Engl. J. Med. https://doi.org/10.1056/NEJMc2119270 (2021).
Cele, S. et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature https://doi.org/10.1038/s41586-021-04387-1 (2021).
Tan, C. W. et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 38, 1073–1078 (2020).
Tan, C. W. et al. Pan-sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N. Engl. J. Med. 385, 1401–1406 (2021).
Zhu, F. et al. WHO international standard for SARS-CoV-2 antibodies to determine markers of protection. Lancet Microbe https://doi.org/10.1016/S2666-5247(21)00307-4 (2021).
Cui, Z. et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 185, 860–871.e13 (2022).
Dejnirattisai, W. et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 185, 467–484.e15 (2022).
Schountz, T., Baker, M. L., Butler, J. & Munster, V. Immunological control of viral infections in bats and the emergence of viruses highly pathogenic to humans. Front. Immunol. 8, 1098 (2017).
Yu, M. et al. Determination and application of immunodominant regions of SARS coronavirus spike and nucleocapsid proteins recognized by sera from different animal species. J. Immunol. Methods 331, 1–12 (2008).
Young, B. E. et al. Comparison of the clinical features, viral shedding and immune response in vaccine breakthrough infection by the Omicron and Delta variants. Preprint at ResearchSquare https://doi.org/10.21203/rs.3.rs-1281925/v1 (2022).
Acknowledgements
We thank Y. Li for helping with coordination of some parts of the study. This work is supported in part by grants from the Singapore National Research Foundation (NRF2016NRF-NSFC002-013, NRF2018NRF-NSFC003SB-002), the National Medical Research Council (STPRG-FY19-001, COVID19RF-003, COVID19RF-060, MOH-000535/MOH-OFYIRG19nov-0002 and OFLCG19May-0034), the Rachadaphiseksomphot Fund (RA(PO)002/64) from the Faculty of Medicine, Chulalongkorn University, the King Chulalongkorn Memorial Hospital Fund for research (HA-64-3300-21-024) and the Biobank, Faculty of Medicine, Chulalongkorn University. A.S. was supported by Bill and Melinda Gates award INV-018944, National Institutes of Health award R01 AI138546 and South African Medical Research Council award 6084-CO-AP-2020.
Author information
Authors and Affiliations
Contributions
C.W.T. and L.-F.W. conceptualized the project. C.W.T., W.N.C., F.Z., A.Y.-Y.Y., B.L.L., W.C.Y., S.C., J.Z., Y.Y.M. and V.C.-W.C. performed the experiments. B.E.Y., N.C., S.-H.H., S.K.MS.P., S.Y.T., W.J., L.K.T., M.I.-C.C., S.W., A.S., O.P. and D.C.L. coordinated cohorts and provided samples used in this study. C.W.T., W.N.C., F.Z., A.Y.-Y.Y., B.L.L. and W.C.Y. performed data analysis. C.W.T., L.-F.W., W.N.C. and F.Z. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
L.-F.W., C.W.T. and W.N.C. are co-inventors of the patented surrogate virus neutralization test marketed by Genscript as cPassTM. The other authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Chris Davis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Amino acid sequence differences between multiple sarbecoviruses.
Phylogenetic trees based on the amino acid sequence of a, RBD and c, Spike were generated using PhyML with Blosum62 model with 1000 bootstrap replicates. Numbers at the branches are percentage bootstrap values for the associated nodes. Scale bar indicates number of substitutions per site. The heat-map for number of amino acid differences among 20 sarbecoviruses on b, RBD and d, Spike, generated from MAFFT alignment with Blosum62 model in Geneious Prime 2022.0.2.
Extended Data Fig. 2 Validation of the 20-plex sVNT with WHO International Standard (IS) SARS-CoV-2 serum panel 20/136.
a, ACE2-RBD binding analysis. 600 beads/RBD were pre-incubated with increased concentration of PE-conjugated ACE2 for 30 min at 37 °C. The Mean Fluorescent Intensity (MFI) were acquired using MagPix Luminex machine. The data presented were from three independent experiments. Error bar indicates standard deviation of mean. b, multiplex sVNT analysis of WHO IS 20/136, with 2-fold serial dilution starting from 1:20. The data presented were from three independent experiments. Error bar indicates standard deviation of mean. c, Calibration of WHO IS unit (IU/ml) using multiplex sVNT on SARS-CoV-2 ancestral strain. Three independent runs were used to perform the correlation of multiplex sVNT inhibition % to IU/ml and logistic regression modelling was performed. The upper and lower 95% confident interval of the mean was plotted in blue shade. The modelled equation for IU/ml to inhibition % and a pseudoR2 (calculated by 1-deviance/null deviance) was marked on the left upper corner. Pearson’s correlation analysis between multiplex sVNT and pVNT on d, ancestral (n = 120) and e, Omicron BA.1 (n = 120). f, Pearson’s correlation analysis between multiplex sVNT and PRNT on Omicron BA.1 (n = 120). Correlation and linear regression analyses were performed in GraphPad Prism 8 using Pearson’s correlation coefficients.
Extended Data Fig. 3 Residues crucial for Nab escape.
Graphical illustration of sarbecoviruses a, spike and b, RBD amino acid sequence alignments. The percentage indicates sequence identity to the SARS-CoV-2 ancestral virus. The N-terminal, receptor binding and S2 domains are shaded in blue. The black line indicates differences to the reference sequence (SARS-CoV-2). The neutralization epitopes for class I-VI nAbs against early pandemic strains (D614 or G614) are derived from published data26,27. c, mutagenesis analysis of SARS-CoV-2 RBD and the NT50 of serum samples derived from individuals who received two (n = 10) and three (n = 10) doses of BNT162b2 and SARS survivors who had received two doses of BNT162b2 (n = 10). The experiment was repeated twice. Data presented was from one experimental replicate. The GMT50 and the fold of reduction were indicated. P value less than 0.05 is considered statistical significance. The exact P values were indicated in the figure. Statistical analysis was performed with two-tailed t-test using GraphPad Prism 8. Each dot denotes the NT50 value of a sample, while the box shows the interquartile range with median at the center, and the whiskers represent the maximum and minimum.
Extended Data Fig. 4 Antigenic map of major variants of SARS-CoV-2 and animal sarbecoviruses.
Antigenic cartography was generated by the neutralization titer 50% (NT50) of 16 sarbecoviruses (color circles) from a, all serum panels examined (n = 394, uncolored squares), b, serum from SARS-vaccinated (n = 11, uncolored squares). The x and y axes represented the antigenic distance, with the space of the grey grid lines showing 1 Antigenic Unit (2-fold dilution in titer).
Extended Data Fig. 5 Neutralization escape of SARS-CoV-2 Omicron sublineages.
pVNT was performed using serum samples derived from individuals received 3 doses of mRNA vaccine (n = 40) against SARS-CoV-2 ancestral, BA.1, BA.2, BA.2 L452R, BA.2 F486V, and BA.5. Number on top of each box plot indicates GMT50. The fold of reduction calculated based on GMT50 of ancestral SARS-CoV-2 against each of the Omicron sublineages is indicated on the graph. Each dot denotes the NT50 value of a sample, while the box shows the interquartile range with median at the center, and the whiskers represent the maximum and minimum.
Supplementary information
Source data
Source Data Fig. 1
Raw data for Fig. 1a,c,d.
Source Data Fig. 2
Raw data for Fig. 2a–d.
Source Data Extended Data Fig. 2
Raw data for Extended Data Fig. 2a–f.
Source Data Extended Data Fig. 3
Raw data for Extended Data Fig. 3c.
Source Data Extended Data Fig. 5
Raw data for Extended Data Fig. 5.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tan, C.W., Chia, W.N., Zhu, F. et al. SARS-CoV-2 Omicron variant emerged under immune selection. Nat Microbiol 7, 1756–1761 (2022). https://doi.org/10.1038/s41564-022-01246-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-022-01246-1
This article is cited by
-
Clinical characteristics and host immunity responses of SARS-CoV-2 Omicron variant BA.2 with deletion of ORF7a, ORF7b and ORF8
Virology Journal (2023)
-
Priming conditions shape breadth of neutralizing antibody responses to sarbecoviruses
Nature Communications (2022)