Abstract
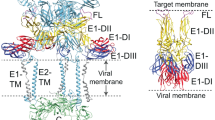
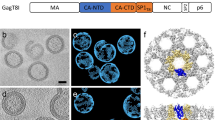
Chikungunya virus (CHIKV) is a representative alphavirus causing debilitating arthritogenic disease in humans. Alphavirus particles assemble into two icosahedral layers: the glycoprotein spike shell embedded in a lipid envelope and the inner nucleocapsid (NC) core. In contrast to matrix-driven assembly of some enveloped viruses, the assembly/budding process of two-layered icosahedral particles remains poorly understood. Here we used cryogenic electron tomography (cryo-ET) to capture snapshots of the CHIKV assembly in infected human cells. Subvolume classification of the snapshots revealed 12 intermediates representing different stages of assembly at the plasma membrane. Further subtomogram average structures ranging from subnanometre to nanometre resolutions show that immature non-icosahedral NCs function as rough scaffolds to trigger icosahedral assembly of the spike lattice, which in turn progressively transforms the underlying NCs into icosahedral cores during budding. Further, analysis of CHIKV-infected cells treated with budding-inhibiting antibodies revealed wider spaces between spikes than in icosahedral spike lattice, suggesting that spacing spikes apart to prevent their lateral interactions prevents the plasma membrane from bending around the NC, thus blocking virus budding. These findings provide the molecular mechanisms for alphavirus assembly and antibody-mediated budding inhibition that provide valuable insights for the development of broad therapeutics targeting the assembly of icosahedral enveloped viruses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Cryo-EM maps reported in this study have been deposited in the Electron Microscopy Data Bank (EMDB) under the following accession codes: EMDB-26446 (released virion), EMDB-26447, −26448, −26449, −26450 (budding intermediates) and EMDB-26451, −26452 (cytosolic NLPs). The publicly deposited atomic model of VEEV TC-83 (PDB:3J0C) was used for comparison to the subtomogram average structure of the CHIKV spike trimer determined in this study. All other data supporting the findings of this study are available within the Article and its supplementary files.
References
Rheinemann, L. & Sundquist, W. I. Virus budding. Encyclopedia of Virology https://doi.org/10.1016/b978-0-12-814515-9.00023-0 (2021).
Brown, R. S., Wan, J. J. & Kielian, M. The alphavirus exit pathway: what we know and what we wish we knew. Viruses 10, 89 (2018).
Silva, L. A. & Dermody, T. S. Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J. Clin. Invest. 127, 737–749 (2017).
Sun, S. et al. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife 2, e00435 (2013).
Tang, J. et al. Molecular links between the E2 envelope glycoprotein and nucleocapsid core in Sindbis virus. J. Mol. Biol. 414, 442–459 (2011).
Zhang, R. et al. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 30, 3854–3863 (2011).
Cheng, R. H. et al. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80, 621–630 (1995).
Jin, J. et al. Neutralizing monoclonal antibodies block chikungunya virus entry and release by targeting an epitope critical to viral pathogenesis. Cell Rep. 13, 2553–2564 (2015).
Fox, J. M. et al. Broadly neutralizing alphavirus antibodies bind an epitope on E2 and inhibit entry and egress. Cell 163, 1095–1107 (2015).
Jin, J. et al. Neutralizing antibodies inhibit chikungunya virus budding at the plasma membrane. Cell Host Microbe 24, 417–428.e5 (2018).
Thaa, B. et al. Differential phosphatidylinositol-3-kinase-akt-mTOR activation by semliki forest and chikungunya viruses is dependent on nsP3 and connected to replication complex internalization. J. Virol. 89, 11420–11437 (2015).
Jose, J., Taylor, A. B. & Kuhn, R. J. Spatial and temporal analysis of alphavirus replication and assembly in mammalian and mosquito cells. mBio 8, e02294–16 (2017).
Chen, L. et al. Implication for alphavirus host-cell entry and assembly indicated by a 3.5Å resolution cryo-EM structure. Nat. Commun. 9, 5326 (2018).
Mukhopadhyay, S., Chipman, P. R., Hong, E. M., Kuhn, R. J. & Rossmann, M. G. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 76, 11128–11132 (2002).
Therkelsen, M. D. et al. Flaviviruses have imperfect icosahedral symmetry. Proc. Natl Acad. Sci. USA 115, 11608–11612 (2018).
Garoff, H., Sjöberg, M. & Cheng, R. H. Budding of alphaviruses. Virus Res. 106, 103–116 (2004).
Soonsawad, P. et al. Structural evidence of glycoprotein assembly in cellular membrane compartments prior to Alphavirus budding. J. Virol. 84, 11145–11151 (2010).
Cogan, D. P. et al. Mapping the catalytic conformations of an assembly-line polyketide synthase module. Science 374, 729–734 (2021).
Bruss, V. & Ganem, D. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl Acad. Sci. USA 88, 1059–1063 (1991).
Ferlenghi, I. et al. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7, 593–602 (2001).
de Haan, C. A., Kuo, L., Masters, P. S., Vennema, H. & Rottier, P. J. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72, 6838–6850 (1998).
Allison, S. L. et al. Two distinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J. Virol. 77, 11357–11366 (2003).
Stange, A., Lüftenegger, D., Reh, J., Weissenhorn, W. & Lindemann, D. Subviral particle release determinants of prototype foamy virus. J. Virol. 82, 9858–9869 (2008).
Wang, P.-G. et al. Efficient assembly and secretion of recombinant subviral particles of the four dengue serotypes using native prM and E proteins. PLoS ONE 4, e8325 (2009).
Heilingloh, C. S. & Krawczyk, A. Role of l-particles during herpes simplex virus infection. Front. Microbiol. 8, 2565 (2017).
Sherer, N. M. & Mothes, W. Cytonemes and tunneling nanotubules in cell–cell communication and viral pathogenesis. Trends Cell Biol. 18, 414–420 (2008).
Nikolic, D. S. et al. HIV-1 activates Cdc42 and induces membrane extensions in immature dendritic cells to facilitate cell-to-cell virus propagation. Blood 118, 4841–4852 (2011).
Sowinski, S. et al. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 10, 211–219 (2008).
Martinez, M. G. & Kielian, M. Intercellular extensions are induced by the alphavirus structural proteins and mediate virus transmission. PLoS Pathog. 12, e1006061 (2016).
Chen, D.-H. et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl Acad. Sci. USA 108, 1355–1360 (2011).
Mukhopadhyay, S., Chipman, P. R., Hong, E. M., Kuhn, R. J. & Rossmann, M. G. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 76, 11128–11132 (2002).
Snyder, J. E. et al. Rescue of infectious particles from preassembled alphavirus nucleocapsid cores. J. Virol. 85, 5773–5781 (2011).
Forsell, K., Xing, L., Kozlovska, T., Cheng, R. H. & Garoff, H. Membrane proteins organize a symmetrical virus. EMBO J. 19, 5081–5091 (2000).
Skoging, U., Vihinen, M., Nilsson, L. & Liljeström, P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure 4, 519–529 (1996).
Jose, J. et al. Interactions of the cytoplasmic domain of Sindbis virus E2 with nucleocapsid cores promote alphavirus budding. J. Virol. 86, 2585–2599 (2012).
Williamson, L. E. et al. Therapeutic alphavirus cross-reactive E1 human antibodies inhibit viral egress. Cell 184, 4430–4446.e22 (2021).
Kim, A. S. et al. Pan-protective anti-alphavirus human antibodies target a conserved E1 protein epitope. Cell 184, 4414–4429.e19 (2021).
Selvarajah, S. et al. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl. Trop. Dis. 7, e2423 (2013).
Mastronarde, D. N. SerialEM: a program for automated tilt series acquisition on tecnai microscopes using prediction of specimen position. Microsc. Microanal. 9, 1182–1183 (2003).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Chen, M. et al. A complete data processing workflow for cryo-ET and subtomogram averaging. Nat. Methods 16, 1161–1168 (2019).
Pettersen, E. F., Goddard, T. D. & Huang, C. C. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. (2004).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
We thank SLAC National Accelerator Laboratory for access and support of these studies, and all SLAC cryo-EM staff for technical support and assistance. We also thank M. Chen for helpful discussions and providing technical advice on data analysis. This research was supported by NIH grants R01AI148382 and S10OD021600 (to W.C.) and R01AI119056 (to G.S.).
Author information
Authors and Affiliations
Contributions
D.C., J.J. and W.C. designed the study. D.C and J.J. performed cryo-EM sample preparation and collected cryo-ET data. D.C. performed 3D reconstruction and subtomogram averaging. D.C., J.J., M.F.S. and W.C. analysed the data. D.C., J.J. and W.C. wrote the manuscript with support from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 RNA replication spherules on the cell surface.
(a) Tomogram slice displaying cell periphery with RNA replication spherules (yellow arrows), budding intermediates (white arrows) and apparently cytosolic NLPs (red arrows). Scale bar: 100 nm. (b) Enlarged tomogram slice of RNA replication spherule with components labeled. Proposed location of the replication complex at the neck of spherules was indicated with a yellow dashed circle. (c) Enlarged slice view of a RNA spherule with a NLP (red dashed circle) in close proximity to the spherule neck (yellow dashed circle), with some thin density (blue arrow) connecting in between. (d) Enlarged slice view of RNA spherule in close proximity to another apparently incompletely-assembled NLP (red dashed circle) and multiple budding particles (white arrows).
Extended Data Fig. 2 Strands of incomplete particles extend from the cell surface.
(a-d) Tomogram slice images display thin extensions of virus budding intermediates in multiple conformations (white dashed boxes, inset images). In all observed cases, beads of linked particles form at the convergences of two membrane surfaces with near-continuous budding intermediates. Scale bars: 200 nm.
Extended Data Fig. 3 CryoET data processing workflow and resolutions of STA structures.
(a) CryoET and data processing workflow for release virions (left), budding intermediates (middle) and budding arrested NLPs (right). For budding intermediates subvolume classification (middle), particles in 5 of the 10 classes (dashed black boxes) from the first multi-reference refinement were subjected to additional classification. 12 distinct intermediate budding conformations (blue boxes) were resolved after two rounds of classification. Note: two maps displayed within a single blue box merged into one class due to overall structure similarity. (b) Gold standard Fourier shell correlation (FSC) plots of subtomogram average structures of budding intermediates and released virions. (c) Slice view of 3D reconstruction of released CHIKV virions. Subnanometer (8.3 Å) resolution of structure is evident by resolved TM helices of E1/E2 in the lipid bilayer. (d) Central section of released icosahedral CHIKV particle reveals density of CP C-protease domain (red) as well as an ordered network of density layer below (purple) that is likely CP N-terminus+gRNA.
Extended Data Fig. 4 Non-icosahedral features of virus particles.
(a) Subtomogram average structures of cytosolic NLPs and NCs from budding-intermediate 3D reconstructions (shown in red). NC structures are overlaid on the density map of the NC from released CHIKV virion (white, transparent). (b) Volta phase plate tomogram slice images of PM with multiple budding intermediates, including late−stage (‘tethered’) particles. Inset is the zoom-in view of the boxed area in (B) displaying relatively absent density at the base of late−stage budding particles (blue arrow). Top of a budding particle, furthest from PM is defined as the leading end, while base of the particle is defined as the trailing end. (c) Subtomogram average structure of ‘tethered intermediate’ shows icosahedral symmetry at the leading end, while trailing end of average shows a disordered final penton and distorted capsomer density in those hexamer units below. Relatively absent density between NC and viral envelope (blue arrows) was also observed at (d) the non-spherical pole of released virions, and (e) the released multi-cored particles. NCs in multi-core particles are labeled with dashed red circles.
Extended Data Fig. 5 Orientation of cytosolic NLPs.
(a) Volta-phase plate tomogram slice image shows budding cell periphery with apparently cytosolic NLPs (red arrows). Scale bar: 50 nm. (b,c) Zoom-in views of NLPs (red arrows). Scale bars: 30 nm. (d) Tomogram slice image of cell with subvolume averages of NLP class II (red, Extended Data Fig. 4) mapped back to the tomogram based on the refined orientation of each particle. 5-fold density consistently oriented towards the PM surface in slices above. (e) Tomogram slice image displays cluster of NLPs (red arrow). Scale bar: 50 nm. (f) In a tomogram slice directly above a single NLP (white box), a penton of spikes (dashed white circle) is identified along with nearby spikes (white arrows). Scale bar: 50 nm.
Extended Data Fig. 6 Different assemblies of spikes on virus-infected cell surface.
(A) Single slice of tomogram depicting virus-induced extension with multiple spike−induced features, including (I) hexameric sheet lattice, (II) budding strand of intermediates, and (III) helical tube. Scale bar 200 nm. (B.I) Tomographic slice view of near-planar lattice (A-I) of trimeric spikes arranged as hexamers (orange) and disrupted region (yellow). (B.II) 3D annotation of spikes (orange) shows disruption of hexameric lattice above NC (A-yellow arrow). (C.I) Tomographic slice view of budding intermediate particle surface (A-II) shows pentamer (yellow) and hexamer (orange) of trimeric spikes. (C.II) 3D annotation of particles on budding strand displays spherical curvature of spike lattice with pentamer (yellow arrow) and hexamer (black arrow). (D.I) Slice view of helical tube (A-III) formed by hexameric lattice of spikes with (D.II) 3D annotation.
Extended Data Fig. 7 Self-assembled structures of spikes alone.
(a-c) Images of CHIKV-infected cell peripheries display cell extensions (white arrows) directly from the cell body. Scale bars: 1 μm. (d & e) Zoom-in images of the boxed regions in B & C, display tubular protein arrays (black arrows) at the terminal end and released vesicle-like assembly products lacking dense cores (white arrows). Scale bars: 200 nm. (f & g) 3D segmentations of cellular features corresponding to (D & E) show the surfaces of tubular extensions contain dense spikes. Segmentations reveal spikes on the surface of tubular membrane extensions that largely exclude bundled cytoskeleton filaments.
Extended Data Fig. 8 Different conformations of spikes before and after assembly in budding shells.
(a,d) Tomogram slice displaying cell periphery with budding intermediates. Scale bars: 200 nm. (b, c, e, f) Enlarged tomogram slices of budding particles. Spike side-views on budding intermediates surface (blue dashed circle) displayed characteristic conformation of spikes assembled in lattice, in contrast to potential individual spikes (yellow arrows) near the base of budding intermediates.
Supplementary information
Supplementary Information
Supplementary Tables 1–2.
Supplementary Video 1
A representative cryo-electron tomogram of a CHIKV-infected U2OS cell.
Supplementary Video 2
A representative cryo-electron tomogram of a NAb C9-treated CHIKV-infected U2OS cell
Rights and permissions
About this article
Cite this article
Chmielewski, D., Schmid, M.F., Simmons, G. et al. Chikungunya virus assembly and budding visualized in situ using cryogenic electron tomography. Nat Microbiol 7, 1270–1279 (2022). https://doi.org/10.1038/s41564-022-01164-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-022-01164-2
This article is cited by
-
Yersinia entomophaga Tc toxin is released by T10SS-dependent lysis of specialized cell subpopulations
Nature Microbiology (2024)
-
Precise targeting for 3D cryo-correlative light and electron microscopy volume imaging of tissues using a FinderTOP
Communications Biology (2023)
-
Chikungunya virus assembly and egress
Nature Microbiology (2022)
-
Snapshots capture self-assembly of dangerous virus
Nature (2022)