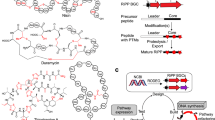
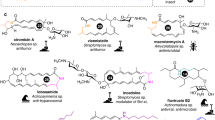
Abstract
The caseinolytic protease (ClpP) is part of a highly conserved proteolytic complex whose disruption can lead to antibacterial activity but for which few specific inhibitors have been discovered. Specialized metabolites produced by bacteria have been shaped by evolution for specific functions, making them a potential source of selective ClpP inhibitors. Here, we describe a target-directed genome mining strategy for discovering ClpP-interacting compounds by searching for biosynthetic gene clusters that contain duplicated copies of ClpP as putative antibiotic resistance genes. We identify a widespread family of ClpP-associated clusters that are known to produce pyrrolizidine alkaloids but whose connection to ClpP has never been made. We show that previously characterized molecules do not affect ClpP function but are shunt metabolites derived from the genuine product of these gene clusters, a reactive covalent ClpP inhibitor. Focusing on one such cryptic gene cluster from Streptomyces cattleya, we identify the relevant inhibitor, which we name clipibicyclene, and show that it potently and selectively inactivates ClpP. Finally, we solve the crystal structure of clipibicyclene-modified Escherichia coli ClpP. Clipibicyclene’s discovery reveals the authentic function of a family of natural products whose specificity for ClpP and abundance in nature illuminate the role of eco-evolutionary forces during bacterial competition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article and its Supplementary Information. The S. cattleya DSM 46488 whole genome sequence is available in GenBank under accession number NC_017586. The structure of ClpPec in complex with clipibicyclene is available in PDB with accession number 7MK5. Source data are provided with this paper.
References
Alexopoulos, J. A., Guarné, A. & Ortega, J. ClpP: a structurally dynamic protease regulated by AAA+ proteins. J. Struct. Biol. 179, 202–210 (2012).
Culp, E. & Wright, G. D. Bacterial proteases, untapped antimicrobial drug targets. J. Antibiot. 70, 366–377 (2017).
Sauer, R. T. & Baker, T. A. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 (2011).
Compton, C. L., Schmitz, K. R., Sauer, R. T. & Sello, J. K. Antibacterial activity of and resistance to small molecule inhibitors of the ClpP peptidase. ACS Chem. Biol. 8, 2669–2677 (2013).
Ju, Y. et al. Discovery of novel peptidomimetic boronate ClpP inhibitors with noncanonical enzyme mechanism as potent virulence blockers in vitro and in vivo. J. Med. Chem. 63, 3104–3119 (2020).
Hackl, M. W. et al. Phenyl esters are potent inhibitors of caseinolytic protease P and reveal a stereogenic switch for deoligomerization. J. Am. Chem. Soc. 137, 8475–8483 (2015).
Böttcher, T. & Sieber, S. A. Structurally refined β-lactones as potent inhibitors of devastating bacterial virulence factors. ChemBioChem 10, 663–666 (2009).
Lakemeyer, M. et al. Tailored peptide phenyl esters block ClpXP proteolysis by an unusual breakdown into a heptamer–hexamer assembly. Angew. Chem. Int. Ed. 58, 7127–7132 (2019).
Li, D. H. S. et al. Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: a model for the ClpX/ClpA-bound state of ClpP. Chem. Biol. 17, 959–969 (2010).
Lee, B. et al. Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat. Struct. Mol. Biol. 17, 471–478 (2010).
Graves, P. R. et al. Mitochondrial protease ClpP is a target for the anticancer compounds ONC201 and related analogues. ACS Chem. Biol. 14, 1020–1029 (2019).
Michel, K. H. & Kastner, R. E. A54556 antibiotics and process for production thereof. US patent application 4492650 A (1985).
Thomy, D. et al. The ADEP biosynthetic gene cluster in Streptomyces hawaiiensis NRRL 15010 reveals an accessory ClpP gene as a novel antibiotic resistance factor. Appl. Environ. Microbiol. 85, e01292-19 (2019).
Sabotič, J. & Kos, J. Microbial and fungal protease inhibitors—current and potential applications. Appl. Microbiol. Biotechnol. 93, 1351 (2012).
Waglechner, N., Culp, E. J. & Wright, G. D. Ancient antibiotics, ancient resistance. EcoSal Plus https://doi.org/10.1128/ecosalplus.ESP-0027-2020 (2021).
Tang, X. et al. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol. 10, 2841–2849 (2015).
Alanjary, M. et al. The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids Res. 45, W42–W48 (2017).
Maxson, T. et al. Targeting reactive carbonyls for identifying natural products and their biosynthetic origins. J. Am. Chem. Soc. 138, 15157 (2016).
Baltz, R. H. Marcel Faber Roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 33, 507–513 (2006).
Navarro-Muñoz, J. C. et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 16, 60–68 (2020).
Schmidt, Y. et al. Biosynthetic origin of the antibiotic cyclocarbamate brabantamide A (SB-253514) in plant-associated Pseudomonas. ChemBioChem 15, 259–266 (2014).
Johnston, C. W., Zvanych, R., Khyzha, N. & Magarvey, N. A. Nonribosomal assembly of natural lipocyclocarbamate lipoprotein-associated phospholipase inhibitors. ChemBioChem 14, 431–435 (2013).
Liu, L. et al. Activation and characterization of bohemamine biosynthetic gene cluster from Streptomyces sp. CB02009. Org. Lett. 22, 4614–4619 (2020).
Hong, Z. et al. Azetidine-containing alkaloids produced by a quorum-sensing regulated nonribosomal peptide synthetase pathway in Pseudomonas aeruginosa. Angew. Chem. 131, 3210–3214 (2019).
Patteson, J. B., Lescallette, A. R. & Li, B. Discovery and biosynthesis of azabicyclene, a conserved nonribosomal peptide in Pseudomonas aeruginosa. Org. Lett. 21, 4955–4959 (2019).
DE, N. et al. Antitumor agents from bohemic acid complex, III. The isolation of marcellomycin, musettamycin, rudolphomycin, mimimycin, collinemycin, alcindoromycin, and bohemamine. J. Nat. Prod. 43, 242–258 (1980).
Bugni, T. S., Woolery, M., Kauffman, C. A., Jensen, P. R. & Fenical, W. Bohemamines from a marine-derived Streptomyces sp. J. Nat. Prod. 69, 1626–1628 (2006).
Viala, J. & Mazodier, P. ClpP-dependent degradation of PopR allows tightly regulated expression of the clpP3 clpP4 operon in Streptomyces lividans. Mol. Microbiol. 44, 633–643 (2002).
Gominet, M., Seghezzi, N. & Mazodier, P. Acyl depsipeptide (ADEP) resistance in Streptomyces. Microbiology 157, 2226–2234 (2011).
Bellier, A., Gominet, M. & Mazodier, P. Post-translational control of the Streptomyces lividans ClgR regulon by ClpP. Microbiology 152, 1021–1027 (2006).
Bellier, A. & Mazodier, P. ClgR, a novel regulator of clp and lon expression in Streptomyces. Society 186, 3238–3248 (2004).
Myronovskyi, M., Welle, E., Fedorenko, V. & Luzhetskyy, A. β-Glucuronidase as a sensitive and versatile reporter in Actinomycetes. Appl. Environ. Microbiol. 77, 5370–5383 (2011).
Nagpal, J. et al. Molecular and structural insights into an asymmetric proteolytic complex (ClpP1P2) from Mycobacterium smegmatis. Sci. Rep. 9, 18019 (2019).
Wang, J., Hartling, J. A. & Flanagan, J. M. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell 91, 447–456 (1997).
Bewley, M. C., Graziano, V., Griffin, K. & Flanagan, J. M. The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes. J. Struct. Biol. 153, 113–128 (2006).
Davies, J., Spiegelman, G. B. & Yim, G. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 9, 445–453 (2006).
Yim, G., Spiegelman, G. B. & Davies, J. E. Separate mechanisms are involved in rifampicin upmodulated and downmodulated gene expression in Salmonella typhimurium. Res. Microbiol. 164, 416–424 (2013).
Vázquez-Laslop, N. & Mankin, A. S. How macrolide antibiotics work. Trends Biochem. Sci. 43, 668–684 (2018).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics (John Innes Foundation, 2000).
Xu, M. et al. GPAHex-A synthetic biology platform for Type IV–V glycopeptide antibiotic production and discovery. Nat. Commun. 11, 5232 (2020).
Yamanaka, K. et al. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl Acad. Sci. USA 111, 1957–1962 (2014).
Luo, Y., Zhang, L., Barton, K. W. & Zhao, H. Systematic identification of a panel of strong constitutive promoters from Streptomyces albus. ACS Synth. Biol. 4, 1001–1010 (2015).
Gietz, R. D. & Schiestl, R. H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007).
Ji, C.-H., Kim, J.-P. & Kang, H.-S. Library of synthetic Streptomyces regulatory sequences for use in promoter engineering of natural product biosynthetic gene clusters. ACS Synth. Biol. 7, 1946–1955 (2018).
Gust, B., Kieser, T. & Chater, K. PCR targeting system in Streptomyces coelicolor A3(2) http://streptomyces.org.uk/redirect/protocol_V1_4.pdf (2002).
Wang, W. et al. An engineered strong promoter for streptomycetes. Appl. Environ. Microbiol. 79, 4484–4492 (2013).
Ahsan, B. Understanding the Activation of Bacterial Protease ClpP by Acyldepsipeptide Antibiotics. MSc thesis, McMaster Univ. (2014).
Marty, M. T. et al. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 87, 4370–4376 (2015).
Cobb, R. E., Wang, Y. & Zhao, H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth. Biol. 4, 723–728 (2015).
Ji, C. H., Kim, H. & Kang, H. S. Synthetic inducible regulatory systems optimized for the modulation of secondary metabolite production in Streptomyces. ACS Synth. Biol. 8, 577–586 (2019).
Culp, E. J. et al. Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics. Nat. Biotechnol. 37, 1149–1154 (2019).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D 67, 293–302 (2011).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Painter, J. & Merritt, E. A. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 (2006).
Acknowledgements
We thank M. Tyers for the gift of pSASS vectors and S. cerevisiae SASy31 and SASy35, W. Houry for the gift of E. coli BL21(DE3) 1146D and pET9a-EcClpP, H. Brötz-Oesterhelt for B. subtilis ΔclpP and V. Larionov for S. cerevisiae VL648N. This research was funded by a Canadian Institutes of Health Research grant (FRN-148463), the Ontario Research Fund and a Canada Research Chair to G.D.W. E.J.C. was supported by a CIHR Vanier Canada Graduate Scholarship. D.S. was supported by a CIHR Postdoctoral Fellowship. Protein structural studies were performed using beamline CMCF-BM at the Canadian Light Source at the University of Saskatchewan, which is supported by the Canada Foundation for Innovation, the Natural Sciences and Engineering Research Council (RGPIN-2018-04968), the National Research Council (NRC), the Canadian Institutes of Health Research, the Government of Saskatchewan and the University of Saskatchewan.
Author information
Authors and Affiliations
Contributions
E.J.C. and G.D.W. conceived the study and designed experiments. A.C.P. and E.J.C. performed target-directed genome mining. G.P. and D.S. performed crystallization, X-ray data collection, processing and model building. D.S. carried out structural analysis, performed peptide mapping, clipibicyclene titration experiments and serine hydrolase profiling. C.H. performed structural elucidation of azabicyclenes and clipibicyclene and purified clipibicyclene. E.J.C. performed all other experiments. E.J.C. and G.D.W. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Herman Sintim, Kim Lewis and Lynn Silver for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Activation of cac, a pyrrolizidine alkaloid-related BGC.
(a) Structures of previously identified bacterial pyrrolizidine alkaloids and related metabolites. (b) RT–PCR results compare the expression of cac in S. cattleya and heterologous producer S. coelicolor M1154 with various engineered constructs. The cluster-situated XRE family transcriptional activator, Cac15, is overexpressed using the plasmid pIJ10257. RT–PCR was performed in biological triplicate and technical duplicate and normalized to the housekeeping sigma factor hrdB. Bars represent the mean with error bars showing standard deviation and dots individual measurements from biological replicates. (c) Schematic shows the design of the refactored construct pCGW-cac-LHK-apra, which becomes pCGW-cac-LHK after removal of the apramycin resistance cassette.
Extended Data Fig. 2 LC–MS/MS of azabicyclenes and ClpP active site peptide.
LC–MS/MS spectra of (a) azabicyclene D, (b) azabicyclene C, and (c) azabicyclene B. Exact mass of predicted ions are shown. MS spectra were acquired in positive mode with 10 eV collision energy. (d) LC–MS/MS spectra ClpP active site peptide that is unmodified (m/z = 855.1, left) or modified (m/z = 964.4 right) with clipibicyclene. Ions corresponding to b and y fragments are labelled. Ions y14-16 missing from the modified peptide’s spectra are predicted to be outside of the mass detector’s range. Inset of the low molecular weight range of the modified peptide shows fragments corresponding to masses observed from azabicyclene B and C.
Extended Data Fig. 3 Proposed biosynthesis of azabicyclenes.
See supplementary discussion for a full description. Coloured carbon atoms track those incorporated by each module of the PKS.
Extended Data Fig. 4 In vivo and in vitro activity of azabicyclenes.
(a) Susceptibility testing of various bacteria species and mammalian cells against azabicyclene C and D (AzaC, AzaD) and bohemamines A and B (BohA, BohB). While azabicyclene D has weak growth inhibitory activity (MIC = 64-128 μg/mL), it is equally active against species where ClpP is dispensable (S. aureus, Bacillus subtilis) and clpP knockout strains (B. subtilis ΔclpP), indicating that its activity is independent of ClpP. ND – not determined. (b) Effect of azabicyclenes on ClpP1P2scatt peptidase activity using (S-LLVY)2 Rhodamine 110 as a substrate (left), and ClpPec proteinase activity using 50 μg/mL FITC-casein as a substrate (right). (c) Effect of bohemamines on ClpP1P2scatt peptidase activity using (S-LLVY)2 Rhodamine 110 as a substrate (left), and ClpPec proteinase activity using 50 μg/mL FITC-casein as a substrate (right). For panels (b) and (c), mean of replicate reactions (n = 2) is shown with error bars representing standard deviation. All experiments were performed on at least two independent occasions with similar results.
Extended Data Fig. 5 Bioactivity of an unstable ClpP inhibitor.
(a) RT–PCR quantification of clpP3 transcripts in the heterologous producer S. coelicolor M1154. Complementation of cac16-17 is provided by the plasmid pIJ-ncac16-17. Mean with error bars showing standard deviation is plotted (n = 3) for three independent cultures. Significance is tested against pCGW with **p = 0.0008 or *p = 0.0392 by two-sided Kruskal-Wallis with Dunn’s multiple comparison test. (b) Extracted ion chromatograms of n-butanolic extracts from S. coelicolor M1154 strains carrying the cac cluster with cac8 deleted, as indicated. Complementation of cac8 is provided by the plasmid pIJ10257-cac8. Results are representative of three independent fermentations. (c) Quantification of results shown in (b) for biological triplicate fermentations (n = 3) with error bars showing standard deviation. (d) S. coelicolor pIJGUS-pClpP3 pSET152 streaked on media containing X-gluc tested against pure azabicyclenes and bohemamines. 40 μg compound is spotted on each disk. NP indicates the bohemamine-type compound NP25302. Cr indicates crude n-butanolic extract of either S. coelicolor M1154 pCGW-cac-LHK (azabicyclene) or S. sp. NBRC110035 (bohemamine) fermentations, where 10 μL is applied to each disk. An agar plug inoculated with S. coelicolor M1154 pCGW-cac-LHK is used as a positive control. (e) S. coelicolor pIJGUS-pClpP3 pSET152 co-streaked with the bohemamine producing organism Streptomyces sp. NBRC110035 or S. coelicolor M1154 pCGW (negative control). (f) Kirby–Bauer growth inhibition assays with agar plugs inoculated with S. coelicolor M1154 strains, as indicted on the left. Indicator strains labelled along the top are environmental Streptomyces spp. (g) Time course of clipibicyclene production using S. coelicolor pIJGUS-pClpP indicator assay. 2.5 μL of each sample is spotted as indicated. All experiments were repeated on at least two independent occasions with similar results.
Extended Data Fig. 6 Reconstitution of ClpP in vitro.
(a) In vitro processing of S. cattleya ClpPs was determined by intact protein MS after incubation alone or with cognate ClpPs, as listed. Incubations were carried out in the presence or absence of ADEP to determine its effect. (b) The activity of a variety of substrates and the effects of activator peptides were measured. % activity is calculated based on rate of the reaction, normalized to that with Suc-LLVY-AMC substrate and no activator peptide. (c) Thermal shift assays of S. cattleya ClpPs in the presence or absence of ADEP. Numbers indicate melting temperature (°C). Assays were performed in technical duplicate with representative curve shown. For panels (a) and (b) means of technical triplicate reactions are reported. All experiments were performed on two independent occasions with similar results.
Extended Data Fig. 7 Behaviour of clipibicyclene in culture supernatant.
(a) Extracted ion chromatogram shows that clipibicyclene (m/z = 347.1605) is removed upon incubation with ClpPec. 85 μM enzyme was incubated with dichloromethane extract of S. coelicolor M1154 pCGW-cac-LHK prior to LC–MS analysis. (b) Stability of clipibicyclene over the course of zero to three days in water or various medias and at the temperatures indicated was assessed by LC–MS quantification.
Extended Data Fig. 8 Structural details of the ClpPec:clipibicyclene axial pore.
(a) Cartoon representation of the asymmetric unit which contains two complete ClpPec tetradecamers. (b) Left, cartoon representation of the axial pore of a ClpPec:clipibicyclene heptamer. The grey circle highlights the unmodelled N-terminal regions of the complex. Protein chains are labelled alphabetically. Right, detailed structure of the N-terminus of chain A (residues 2–20) depicted in stick representation. The 2mFo-DFc is shown in black mesh and contoured at 1σ. (c) Superposition of the ClpP:clipibicyclene tetradecamer (left) and the monomer (right) with apo ClpP, coloured teal and white, respectively. The apo ClpPec model was obtained from the PDB (1TYF).
Extended Data Fig. 9 Structures of the clipibicyclene adduct in from each chain.
The clipibicyclene adduct is depicted in stick representation and is coloured by atom (C, white; N, blue; O, red). The adducts were defined by a carbamoyl linkage between the Oɣ of Ser97 and the nitrogen of the azetine ring (Fig. 6a). As expected, the seven-membered ring of clipibicyclene is no longer intact. Instead, it is replaced by an imide moiety connecting the azetine portion of the adduct to its aliphatic tail. The 2mFo-DFc map is shown in black mesh and contoured at 0.9 σ and the Fo-Fc omit map is coloured green mesh and contoured at 3 σ. The chain to which each adduct is linked is labelled alphabetically. We use the adduct from chain A as the representative conformer to describe the ClpPec:clipibicyclene complex.
Supplementary information
Supplementary Information
Supplementary Discussion, Note, Figs. 1–5 and references.
Supplementary Tables
Supplementary Tables 1–13.
Source data
Source Data Fig. 3
Excel file, raw data for 3a,b.
Source Data Fig. 4
Unprocessed gel, 4c Excel file.
Source Data Fig. 4
Raw data for 4a,b.
Source Data Fig. 5
Excel file, raw data for 5c,f.
Source Data Extended Data Fig. 1
Excel file with raw data for 1b.
Source Data Extended Data Fig. 4
Excel file with raw data for 4b,c.
Source Data Extended Data Fig. 5
Excel file with raw data for 5a,c.
Rights and permissions
About this article
Cite this article
Culp, E.J., Sychantha, D., Hobson, C. et al. ClpP inhibitors are produced by a widespread family of bacterial gene clusters. Nat Microbiol 7, 451–462 (2022). https://doi.org/10.1038/s41564-022-01073-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-022-01073-4