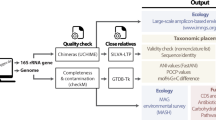
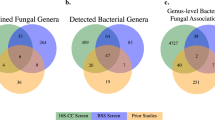
Abstract
The identification and proper naming of microfungi, in particular plant, animal and human pathogens, remains challenging. Molecular identification is becoming the default approach for many fungal groups, and environmental metabarcoding is contributing an increasing amount of sequence data documenting fungal diversity on a global scale. This includes lineages represented only by sequence data. At present, these taxa cannot be formally described under the current nomenclature rules. By considering approaches used in bacterial taxonomy, we propose solutions for the nomenclature of taxa known only from sequences to facilitate consistent reporting and communication in the literature and public sequence repositories.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
27 May 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41564-021-00921-z
References
Richards, T. A., Leonard, G. & Wideman, J. G. What defines the "kingdom" Fungi? Microbiol. Spectr. 5, 57–77 (2017).
Hawksworth, D. L. & Lücking, R. in The Fungal Kingdom (eds Heitman, J. et al.) 79–95 (ASM Press, 2017).
Berbee, M. L., James, T. Y. & Strullu-Derrien, C. Early diverging fungi: diversity and impact at the dawn of terrestrial life. Annu. Rev. Microbiol. 71, 41–60 (2017).
Lücking, R. et al. Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal barcoding? IMA Fungus 11, 14 (2020).
Wijayawardene, N. N. et al. Outline of Fungi and fungus-like taxa. Mycosphere 11, 1060–1456 (2020).
Beakes, G. W. & Thines, M. in Handbook of the Protists 2nd edn (eds Archibald, J. M. et al.) 435–505 (Springer, 2017).
Turland, N. J. et al. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) 2018 (Koeltz Botanical Books, 2018).
Hawksworth, D. L. et al. The Amsterdam declaration on fungal nomenclature. IMA Fungus 2, 105–112 (2011).
Schoch, C. L. et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl Acad. Sci. USA 109, 6241–6246 (2012).
Forsberg, K. et al. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 57, 1–12 (2019).
Daniel, H.-M., Lachance, M.-A. & Kurtzman, C. P. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie van Leeuwenhoek 106, 67–84 (2014).
Shen, X. X. et al. Tempo and mode of genome evolution in the budding yeast subphylum. Cell 175, 1533–1545 (2018).
Rush, T. A. et al. Variation in the internal transcribed spacer region of Phakopsora pachyrhizi and implications for molecular diagnostic assays. Plant Dis. 103, 2237–2245 (2019).
Zhang, N. et al. Generic names in Magnaporthales. IMA Fungus 7, 155–159 (2016).
Cogliati, M. et al. Genotypes and population genetics of Cryptococcus neoformans and Cryptococcus gattii species complexes in Europe and the Mediterranean area. Fungal Genet. Biol. 129, 16–29 (2019).
Firacative, C., Trilles, L. & Meyer, W. MALDI–TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PLoS ONE 7, e37566 (2012).
Liu, X. Z. et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Stud. Mycol. 81, 85–147 (2015).
Nguyen, N. H. et al. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248 (2016).
Maryani, N. et al. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud. Mycol. 92, 155–194 (2019).
Liu, F., Wang, M., Damm, U., Crous, P. W. & Cai, L. Species boundaries in plant pathogenic fungi: a Colletotrichum case study. BMC Evol. Biol. 16, 81–81 (2016).
Thines, M., Telle, S., Ploch, S. & Runge, F. Identity of the downy mildew pathogens of basil, coleus, and sage with implications for quarantine measures. Mycol. Res. 113, 532–540 (2009).
Ruppert, K. M., Kline, R. J. & Rahman, M. S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 17, e00547 (2019).
James, T. Y., Stajich, J. E., Hittinger, C. T. & Rokas, A. Toward a fully resolved fungal tree of life. Annu. Rev. Microbiol. 74, 291–313 (2020).
Jones, M. D. M. et al. Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474, 200–203 (2011).
Rosling, A. et al. Archaeorhizomycetes: unearthing an ancient class of ubiquitous soil Fungi. Science 333, 876–879 (2011).
Page, R. D. DNA barcoding and taxonomy: dark taxa and dark texts. Philos. Trans. R. Soc. B 371, 20150334 (2016).
Sayers, E. W. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. D1, D9–D16 (2020).
Anslan, S. et al. Great differences in performance and outcome of high-throughput sequencing data analysis platforms for fungal metabarcoding. MycoKeys 39, 29–40 (2018).
Nilsson, R. H. et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264 (2019).
Öpik, M. et al. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol. 188, 223–241 (2010).
Schoch, C. L. et al. NCBI taxonomy: a comprehensive update on curation, resources and tools. Database 2020, baaa062 (2020).
Parker, C. T., Tindall, B. J. & Garrity, G. M. International Code of Nomenclature of Prokaryotes. Prokaryotic Code (2008 revision). Int. J. Syst. Evol. Microbiol. 69, S7–S111 (2019).
Lücking, R. & Hawksworth, D. L. Formal description of sequence-based voucherless Fungi: promises and pitfalls, and how to resolve them. IMA Fungus 9, 143–166 (2018).
Thines, M. et al. Ten reasons why a sequence-based nomenclature is not useful for fungi anytime soon. IMA Fungus 9, 177–183 (2018).
Torres-Cruz, T. J. et al. Bifiguratus adelaidae, gen. et sp nov., a new member of Mucoromycotina in endophytic and soil-dwelling habitats. Mycologia 109, 363–378 (2017).
Imachi, H. et al. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature 577, 519–525 (2020).
Overmann, J. Significance and future role of microbial resource centers. Syst. Appl. Microbiol. 38, 258–265 (2015).
Parte, A. C., Sarda Carbasse, J., Meier-Kolthoff, J. P., Reimer, L. C. & Goker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612 (2020).
Louca, S., Mazel, F., Doebeli, M. & Parfrey, L. W. A census-based estimate of Earth’s bacterial and archaeal diversity. PLoS Biol. 17, e3000106 (2019).
Murray, A. E. et al. Roadmap for naming uncultivated Archaea and Bacteria. Nat. Microbiol. 5, 987–994 (2020).
Ryberg, M. & Nilsson, R. H. New light on names and naming of dark taxa. MycoKeys 30, 31–39 (2018).
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
de Beer, Z. W. et al. Hawksworthiomyces gen. nov. (Ophiostomatales), illustrates the urgency for a decision on how to name novel taxa known only from environmental nucleic acid sequences (ENAS). Fungal Biol. 120, 1323–1340 (2016).
Khan, F. et al. Naming the untouchable—environmental sequences and niche partitioning as taxonomical evidence in fungi. IMA Fungus 11, 23 (2020).
Lücking, R. & Moncada, B. Dismantling Marchandiomphalina into Agonimia (Verrucariaceae) and Lawreymyces gen. nov. (Corticiaceae): setting a precedent to the formal recognition of thousands of voucherless fungi based on type sequences. Fungal Divers. 84, 119–138 (2017).
Kirk, P. M. Nomenclatural novelties. Index Fungi 1, 1 (2012).
May, T. W., Redhead, S. A., Lombard, L. & Rossman, A. Y. XI International Mycological Congress: report of Congress action on nomenclature proposals relating to fungi. IMA Fungus 9, xxii–xxvii (2018).
Větrovský, T. et al. GlobalFungi: global database of fungal records from high-throughput-sequencing metabarcoding studies. Sci. Data 7, 228 (2020).
Parks, D. H. et al. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 38, 1098–1098 (2020).
Lee, M. R., & Hawkes, C. V. Plant and soil drivers of whole-plant microbiomes: variation in switchgrass fungi from coastal to mountain sites. Phytobiomes J. https://doi.org/10.1094/PBIOMES-07-20-0056-FI (2020).
Tang, J., Liu, J., Zhang, M. Y. & Mei, Q. Visualizing large-scale and high-dimensional data. In WWW ‘16: Proc. 25th International Conference on World Wide Web 287–297 (International World Wide Web Conferences Steering Committee, 2016); https://doi.org/10.1145/2872427.2883041
Vu, D., Groenewald, M. & Verkley, G. Convolutional neural networks improve fungal classification. Sci. Rep. 10, 12628 (2020).
Acknowledgements
Work by C.L.S. and B.R. was supported by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health in Bethesda, Maryland, USA. D.M.G. received support through the National Science Foundation (NSF) grant DEB-1655980 and Project 4655 of the Pennsylvania State Agricultural Experiment Station. E.M. acknowledges CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil) and FACEPE (Fundação de Amparo à Ciência e Tecnologia de Pernambuco, Brazil). K.D.H. thanks the Thailand Research Fund, grant RDG6130001, entitled “Impact of Climate Change on Fungal Diversity and Biogeography in the Greater Mekong Subregion”. The USDA Hatch project 1010662 is acknowledged for support to M.C.A. M.Ö. was supported by the European Regional Development Fund (Centre of Excellence EcolChange). M.T. acknowledges LOEWE for funding in the framework of the Centre for Translational Biodiversity Genomics (TBG) and the German Science Foundation. N.Z. acknowledges the NSF of the United States (DEB-1452971). P.R.J. was supported through the Manaaki Whenua Biota Portfolio with funding from the Science and Innovation Group of the New Zealand Ministry of Business, Innovation and Employment. R.J. thanks the University of Mauritius for research support. We thank S. Redhead for nomenclatural advice. R. Sanders provided the update for the fungal ITS data in the SRA.
Author information
Authors and Affiliations
Contributions
The present manuscript was first discussed among members of the ICTF. Based on contributions from this initial discussion, a first version of the manuscript was drafted by R.L., C.L.S., M.C.A., B.R. and A.N.M. This version was distributed among the ICTF and to selected colleagues outside the ICTF, and all comments were recorded and incorporated. Based on these initial comments, lead and co-authorship was determined, additional co-authors then including T.A., H.A.A., G.C., P.W.C., I.S.D., D.M.G., D.L.H., K.D.H., L.I., R.J., P.R.J., P.M.K., E.M., T.W.M., W.M., M.Ö., V.R., M.S., M.T., D.V., A.M.Y. and N.Z. The revised draft was circulated two more times among all authors for additional comments before submission. After the first review, H.R.N. was invited as an additional co-author to provide specific input regarding dark taxa and the role of the UNITE database in the proposed alternatives for dark taxa nomenclature. Apart from contributing generally to the manuscript, D.L.H. and T.W.M. revised the nomenclatural details included in Box 1 and Fig. 1. B.R. and D.V. also assisted in technical aspects regarding the SRA and Fig. 3. P.W.C., R.L., W.M., M.T. and A.M.Y. organized the photographs used in Fig. 2 from their working groups. The final draft was approved by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Microbiology thanks Michaela Lackner, Emma Steenkamp and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
References for the timeline events depicted in Fig. 2.
Rights and permissions
About this article
Cite this article
Lücking, R., Aime, M.C., Robbertse, B. et al. Fungal taxonomy and sequence-based nomenclature. Nat Microbiol 6, 540–548 (2021). https://doi.org/10.1038/s41564-021-00888-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-021-00888-x
This article is cited by
-
Singleton-based species names and fungal rarity: Does the number really matter?
IMA Fungus (2024)
-
The yeast genus Tardiomyces gen. nov. with one new species and two new combinations
Infection (2024)
-
Genomic characterization and radiation tolerance of Naganishia kalamii sp. nov. and Cystobasidium onofrii sp. nov. from Mars 2020 mission assembly facilities
IMA Fungus (2023)
-
MycoNews 2022: editorial, news, reports, awards, personalia, and book news
IMA Fungus (2023)
-
Richer than Gold: the fungal biodiversity of Reserva Los Cedros, a threatened Andean cloud forest
Botanical Studies (2023)