Abstract
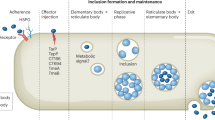
Obligate intracellular bacteria such as Chlamydia trachomatis undergo a complex developmental cycle between infectious, non-replicative elementary-body and non-infectious, replicative reticulate-body forms. Elementary bodies transform to reticulate bodies shortly after entering a host cell, a crucial process in infection, initiating chlamydial replication. As Chlamydia fail to replicate outside the host cell, it is unknown how the replicative part of the developmental cycle is initiated. Here we show, using a cell-free approach in axenic media, that the uptake of glutamine by the bacteria is crucial for peptidoglycan synthesis, which has a role in Chlamydia replication. The increased requirement for glutamine in infected cells is satisfied by reprogramming the glutamine metabolism in a c-Myc-dependent manner. Glutamine is effectively taken up by the glutamine transporter SLC1A5 and metabolized via glutaminase. Interference with this metabolic reprogramming limits the growth of Chlamydia. Intriguingly, Chlamydia failed to produce progeny in SLC1A5-knockout organoids and mice. Thus, we report on the central role of glutamine for the development of an obligate intracellular pathogenic bacterium and the reprogramming of host glutamine metabolism, which may provide a basis for innovative anti-infection strategies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Microarray data have been deposited in the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) of the National Center for Biotechnology Information and can be accessed with the GEO accession numbers GSE147538 and GSE147539. Any other data that support the findings of this study are available from the corresponding author on request. Source data are provided with this paper.
Code availability
The code described here has been integrated into the Galaxy codebase and released under the Academic Free License (AFL) v. 3.0 (https://github.com/galaxyproject/galaxy). A free public version of Galaxy can be accessed at https://usegalaxy.org. For the transcriptomics analysis on sequencing data deposited at the GEO (GSE147538), open access code implemented in bowtie, samtools, edgeR and GSEA tool and packages was utilized.
Change history
15 February 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41564-021-00874-3
References
Newman, L. et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS ONE 10, e0143304 (2015).
Liechti, G. W. et al. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–510 (2014).
Liechti, G. et al. Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog. 12, e1005590 (2016).
Stephens, R. S. et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759 (1998).
Iliffe-Lee, E. R. & McClarty, G. Glucose metabolism in Chlamydia trachomatis: the ‘energy parasite’ hypothesis revisited. Mol. Microbiol. 33, 177–187 (1999).
Mehlitz, A. et al. Metabolic adaptation of Chlamydia trachomatis to mammalian host cells. Mol. Microbiol. 103, 1004–1019 (2017).
Omsland, A., Sager, J., Nair, V., Sturdevant, D. E. & Hackstadt, T. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc. Natl Acad. Sci. USA 109, 19781–19785 (2012).
Kubo, A. & Stephens, R. S. Substrate-specific diffusion of select dicarboxylates through Chlamydia trachomatis PorB. Microbiology 147, 3135–3140 (2001).
Belland, R. J. et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl Acad. Sci. USA 100, 8478–8483 (2003).
Belland, R. J. et al. Transcriptome analysis of chlamydial growth during IFN-γ-mediated persistence and reactivation. Proc. Natl Acad. Sci. USA 100, 15971–15976 (2003).
Miller, D. M., Thomas, S. D., Islam, A., Muench, D. & Sedoris, K. c-Myc and cancer metabolism. Clin. Cancer Res. 18, 5546–5553 (2012).
Gao, P. et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 (2009).
Wise, D. R. et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl Acad. Sci. USA 105, 18782–18787 (2008).
Al-Zeer, M. A. et al. Chlamydia trachomatis prevents apoptosis via activation of PDPK1–MYC and enhanced mitochondrial binding of hexokinase II. EBioMedicine 23, 100–110 (2017).
Welcker, M. et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl Acad. Sci. USA 101, 9085–9090 (2004).
Sears, R. C. The life cycle of C-myc: from synthesis to degradation. Cell Cycle 3, 1133–1137 (2004).
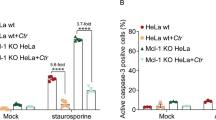
Rajalingam, K. et al. Mcl-1 is a key regulator of apoptosis resistance in Chlamydia trachomatis-infected cells. PLoS ONE 3, e3102 (2008).
Subbarayal, P. et al. EphrinA2 receptor (EphA2) is an invasion and intracellular signaling receptor for Chlamydia trachomatis. PLoS Pathog. 11, e1004846 (2015).
Patel, A. L. et al. Activation of epidermal growth factor receptor is required for Chlamydia trachomatis development. BMC Microbiol. 14, 277 (2014).
Adhikary, S. & Eilers, M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6, 635–645 (2005).
Frame, S., Cohen, P. & Biondi, R. M. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell 7, 1321–1327 (2001).
Lorenzin, F. et al. Different promoter affinities account for specificity in MYC-dependent gene regulation. eLife 5, e15161 (2016).
Cortina, M. E., Ende, R. J., Bishop, R. C., Bayne, C. & Derre, I. Chlamydia trachomatis and Chlamydia muridarum spectinomycin resistant vectors and a transcriptional fluorescent reporter to monitor conversion from replicative to infectious bacteria. PLoS ONE 14, e0217753 (2019).
Dejure, F. R. et al. The MYC mRNA 3′-UTR couples RNA polymerase II function to glutamine and ribonucleotide levels. EMBO J. 36, 1854–1868 (2017).
Kekuda, R. et al. Cloning of the sodium-dependent, broad-scope, neutral amino acid transporter Bo from a human placental choriocarcinoma cell line. J. Biol. Chem. 271, 18657–18661 (1996).
van Geldermalsen, M. et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene 35, 3201–3208 (2016).
Masle-Farquhar, E., Broer, A., Yabas, M., Enders, A. & Broer, S. ASCT2 (SLC1A5)-deficient mice have normal B-cell development, proliferation, and antibody production. Front. Immunol. 8, 549 (2017).
Kermack, A. J. et al. Amino acid composition of human uterine fluid: association with age, lifestyle and gynaecological pathology. Hum. Reprod. 30, 917–924 (2015).
Behjousiar, A., Kontoravdi, C. & Polizzi, K. M. In situ monitoring of intracellular glucose and glutamine in CHO cell culture. PLoS ONE 7, e34512 (2012).
Allan, I. & Pearce, J. H. Differential amino acid utilization by Chlamydia psittaci (strain guinea pig inclusion conjunctivitis) and its regulatory effect on chlamydial growth. J. Gen. Microbiol. 129, 1991–2000 (1983).
Allan, I. & Pearce, J. H. Amino acid requirements of strains of Chlamydia trachomatis and C. psittaci growing in McCoy cells: relationship with clinical syndrome and host origin. J. Gen. Microbiol. 129, 2001–2007 (1983).
Jin, L., Alesi, G. N. & Kang, S. Glutaminolysis as a target for cancer therapy. Oncogene 35, 3619–3625 (2016).
Porcheray, F. et al. Glutamate metabolism in HIV-infected macrophages: implications for the CNS. Am. J. Physiol. Cell Physiol. 291, C618–C626 (2006).
Chambers, J. W., Maguire, T. G. & Alwine, J. C. Glutamine metabolism is essential for human cytomegalovirus infection. J. Virol. 84, 1867–1873 (2010).
Fontaine, K. A., Camarda, R. & Lagunoff, M. Vaccinia virus requires glutamine but not glucose for efficient replication. J. Virol. 88, 4366–4374 (2014).
Thai, M. et al. Adenovirus E4ORF1-induced MYC activation promotes host cell anabolic glucose metabolism and virus replication. Cell Metab. 19, 694–701 (2014).
Thai, M. et al. MYC-induced reprogramming of glutamine catabolism supports optimal virus replication. Nat. Commun. 6, 8873 (2015).
Sanchez, E. L., Carroll, P. A., Thalhofer, A. B. & Lagunoff, M. Latent KSHV infected endothelial cells are glutamine addicted and require glutaminolysis for survival. PLoS Pathog. 11, e1005052 (2015).
Mall, A. et al. Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science 359, 563–567 (2018).
Lee, W. N., Byerley, L. O., Bergner, E. A. & Edmond, J. Mass isotopomer analysis: theoretical and practical considerations. Biol. Mass Spectrom. 20, 451–458 (1991).
Karunakaran, K., Subbarayal, P., Vollmuth, N. & Rudel, T. Chlamydia-infected cells shed Gp96 to prevent chlamydial re-infection. Mol. Microbiol. 98, 694–711 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Kessler, M. et al. Chronic Chlamydia infection in human organoids increases stemness and promotes age-dependent CpG methylation. Nat. Commun. 10, 1194 (2019).
Acknowledgements
We thank S. Broer and A. Enders (Australian National University) for providing the Slc1a5-knockout mice. We thank N. Kurmasheva for supporting the organoid experiment. We thank C. Gehring and D. Bunsen for processing the transmission electron microscopy samples. We thank I. Derre for the EB–RB reporter strain Ct mCh(GroL2) GFP(OmcAL2), V. Kozjak-Pavlovic for the cell line expressing shRNA against GLS1, and A. Demuth and W. Goebel for critically reading the manuscript. K.R. was partially funded by the Frauenbüro in the frame of the ‘Qualification for junior scientist for professorship programme’ and the Department of Biomedicine, Aarhus University. This research work was supported by the German Research Foundation (DFG; grant nos. WO 2108/1–1 (to W.E.), SCHU 2670/1–1 (to A.S.) and GRK 2157 ‘3D-Infect’ (to T.R.), and the European Research Council (grant no. ERC-2018-ADG/NCI-CAD to T.R.). A.B. was supported by grants from the German Excellence Initiative to the Graduate School of Life Sciences.
Author information
Authors and Affiliations
Contributions
K.R. and T.R. designed the experiments. The experiments were performed by K.R., N.V. and T.F.W. Next-generation sequencing on an Illumina platform was performed by A.B. and E.W., and the resulting RNA-seq data were analysed by R.S. and K.R. The samples for mass spectrometry were prepared by K.R. and S.J.-R. N.V., C.H., M.S. and W.E. performed the metabolic flux analysis of axenic cultures and data interpretation. S.J.-R., W.S. and A.S. performed metabolic flux analysis in host cell cultures. F.R.D. and M.E. provided plasmids and cell lines. Click reagents were synthesized by J.F. and J.S. K.R. and T.R. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Glutamine metabolism in Chlamydia.
a. Chlamydial EBs were purified and incubated in axenic media containing G6P with or without Gln. After 2 h NADH levels were measured as described in the Method section. Data is represented Scatter plot with bars overlaid with data points as dot plots. Error bar was defined as mean with SEM. One-way ANOVA was used to analyse the p value, F = 33.88, Df=8. p value ** =0.0012, p value ** =0.014. n = 3. b. Scheme showing the shuttling of 13C and 15N flux from U-[13C5] Gln and 15N (amine) Gln from the axenic culture into Chlamydia for DAP biosynthesis. *Alanine highlights its use in the DAP biosynthesis (shown at the lower part of the cartoon). Red circles indicate M + 5, yellow M + 4 and green M + 3. c. Detailed analysis of DAP: The following raw data show mass traces and mass spectra from the [13C5] Gln culture after sample preparation with acid hydrolysis. The 24 h sample shows a clear 13C incorporation pattern whereas the 0 h sample shows only natural 13C ratios. These data are derived from three independent experiments d. Renografin-purified EBs were incubated in axenic medium with 15N-labelled glutamine specimens and analysed via GC-MS (see Methods for details). The data is derived from three independent experiments. e. Chlamydial EBs were purified using renografin gradient (Input) and incubated in axenic culture containing either only G6P or G6P together with Gln. After 2 or 6 h of incubation, the bacteria were pelleted and analysed by transmission electron microscopy. Representative images from three independent experiments are shown. Scale bar = 2 µM. f. Renografin-purified EBs were incubated with either G6P or G6P and glutamine for 2 h. The bacteria were then used to infect freshly plated HeLa cells. 24 hpi, the inclusion forming units were calculated and plotted in the graph. The number of inclusions in G6P and glutamine-treated sample was normalized against G6P-treated sample. Data is represented Scatter plot with bars overlaid with data points as dot plots. Error bar was defined as mean with SEM. Paired Student’s t-test (two-tailed) was used to analyse the p value, t = 29, Df=2. p value ** =0.0012, n = 3. g. Forward versus side scatter (FSC vs SSC) gating for the FACS analysis presented in Fig. 1g. Signals in yellow represent chlamydial particles and in magenta the debris from the cells. The data represents three independent observation in FACS. h. Renografin-purified EBs were incubated in axenic media containing only G6P or G6P together with Gln. After 12 h the bacteria were pelleted and total RNA was isolated for RNA-seq (see Methods). Heat map was created on the whole gene regulation. The numbers on the left indicate the Chlamydia gene numbers. For details see deposited data set GSE147539. i. Renografin-purified EBs were incubated in axenic media containing only G6P or G6P together with Gln. After 2, 6 and 12 h the bacteria were pelleted and total RNA was isolated to perform qPCR of selected early genes. The values were normalized to the RNA levels of the G6P samples. n = 2. Error bar was defined as mean with SEM.
Extended Data Fig. 2 Chlamydial infection triggers altered glutamine metabolism in infected cells.
Diagram outlining the differential use of glutamine (Gln) by the metabolism of host and chlamydial cells. Host cells feed Gln-derived α-ketoglutarate (α-KG) into their complete TCA cycle. This leads to the production of M + 4, M + 2 and M + 1 isotopologues of all TCA cycle intermediates generated by the first, second and third round of the cycle, respectively. Alternatively, host cells can also use reductive carboxylation of Gln-derived α-KG to generate M + 5 citrate (Cit), which is further converted to oxaloacetate (Oxa) by the host cell metabolism. The different isotopologues of Oxa are then used to generate M + 4, M + 3, M + 2 and M + 1 labelled aspartate (Asp). As no Gln-derived labelling of pyruvate was observed in infected cells, it can be concluded that the gluconeogenesis enzymes required for the conversion of oxaloacetate to pyruvate are inactive. In contrast, the truncated TCA cycle of Chlamydia can only produce M + 4 isotopologues of alpha-KG, succinate (Suc), fumarate (Fum), malate (Mal) and Oxa, resulting in M + 4 labelled Asp. Infected cells increase the uptake of Gln from the culture media and enhance its catabolism to provide metabolic intermediates for bacterial growth.
Extended Data Figure. 3 Signalling pathways involved in the stabilization of c-Myc in infected cells.
a. HeLa229 were left uninfected or infected with Chlamydia at an MOI of 1 for 24 and 36 h. RNA was isolated and qPCR was performed to analyse the levels of c-Myc target genes (n = 3). Error bar was defined as mean with SEM. b/c/d. Human fimb/HUVEC (b), mouse fimb (c) and U2OS cells (d) were infected with C. trachomatis serovar L2 at an MOI of 1 for various time points. The cells were lysed and analysed for c-Myc expression via Western blot. ß Actin serve as the loading control and cHsp60 indicates the infection level. n = 3. e–g. HeLa229 cells were infected with either C. trachomatis serovar D (e), C. pneumoniae (f) or C. muridarum (g) for different time points as indicated and the cell lysate were analysed for c-Myc expression by Western blot. ß Actin serves as the loading control and cHsp60 indicates the infection level. n = 3. h. HeLa229 cells were infected with Chlamydia at an MOI of 1 for different time points. Total RNA was isolated and levels of c-Myc mRNA were detected using qPCR. Data is represented Scatter plot with bars overlaid with data points as dot plots. Error bar was defined as mean with SEM. One-way ANOVA with Tukeys multiple comarison was used for analysis. p value **** <0.0001 ****; p value*** =0.0002, F = 82.5, Df=14, n = 3. i. HUVEC cells were infected with Chlamydia for 24 hpi, the cells were fixed with PFA and immunostained for c-Myc (green), cHsp60 (red) and phalloidin (blue). All experiments were done in three technical replicates. j. HeLa229 cells were either left uninfected (UI) or infected with Chlamydia at an MOI of 1 for 24 or 36 hpi. The cells were lysed and the cytoplasmic and nuclear fraction was isolated as explained in the Methods section. The lysate was analysed using Western blotting for c-Myc. GAPDH was used as a control for cytoplasmic fraction and Histone for nuclear fraction. The data represent three independent experiment.
Extended Data Fig. 4 c-Myc is downstream of PI3K/MAPK pathway and critical for chlamydial growth.
U2OS cells were treated with different concentrations of anhydrous tetracycline (AHT) to investigate adverse effects of AHT on chlamydial infection. The cells were then infected with Chlamydia at an MOI 1 for 30 h. The cells were lysed and analysed by Western blotting for cHsp60 (infection level) and ß Actin (loading control). UI, uninfected; n = 3. b. Infectivity assay from the experiment shown in Fig. 4a,b. AHT-induced and/or U0126/Ly294002 cells were infected with Chlamydia for 30 h and the cells were lysed with glass beads. Different dilutions of the lysate were used to infect freshly plated HeLa229 cells. After 24 h the cells were lysed and the infection level (cHsp60) was detected by Western blotting. ß Actin serve as the loading control. The data represent three independent experiment. n = 3. c. The number of inclusions from the above experiment was counted to plot the graph presented in scatter box plot overlaid with data point overlaid in the graph. p value was determined by one-way ANOVA with Tukey’s multiple comparisons test (F = 19.75, Df=12). Error bar was defined as mean with SEM. n = 3. * indicates p value <0.05 and ** indicates p value <0.01. d. HeLa229 cells were treated with the chemical c-Myc inhibitor 10058-F4. The cells were infected with Chlamydia for 36 h and then lysed to infect freshly plated HeLa cells. The number of cells and number of inclusions were counted to plot the graph (box and whisker plot with a minimum and maximum range of values overlaid in the graph). n = 3. Paired t test (two tailed) was used for analysis p value <0.001 indicates **, t = 4.831, df=2. e. HeLa229 cells carrying an inducible shRNA expression cassette to silence c-Myc expression were either induced with AHT or left uninduced. The cells were infected with Chlamydia at an MOI 1 for 24 h. The cells were lysed and analysed using Western blotting. cHsp60 indicated the infection level, ß Actin served as loading control. n = 3. f. A siRNA pool was used to knock down c-Myc in HeLa cells. After 48 h of transfection, the cells were infected with Chlamydia for 24 h and then lysed to infect HeLa cells. The number of cells and number of inclusions were counted to plot the graph (box and whisker plot with a minimum and maximum range of values overlaid in the graph). n = 3. Paired t test (two tailed) was used for analysis p value <0.001 indicates **, t = 7.853, df=2.
Extended Data Fig. 5 Glutamine is a limiting metabolite for chlamydial intracellular growth.
a–c. Plots of gene set enrichment analyses (GSEA) performed with RNA-seq data of the gene set involved in „amino-acid transport and metabolism“. Gene expression profiles of cells infected for 12, 24 and 36 h and uninfected cells were analysed by RNA-seq. Gene expression changes between infected and uninfected cells were compared by GSEA to all gene sets in the MSigDB C5 (GO Term) collection and a selected gene set is shown. Vertical black bars indicate the position of genes in the „positive regulation of amino acid transporters and amino acid/cellular amine metabolism“ and the enrichment score is shown as a green line. (NES: normalized enrichment score). For statistical details see Method section. The data represents three independent experiments. d. HeLa229 cells were grown in in basic formulation of DMEM containing 4.5 g/l D-glucose and incubated with different concentrations of glutamine and infected with Chlamydia at an MOI 1 for 30 h. The cells were lysed and analysed by western blotting. cHsp60 indicates the infection level ß Actin serve as the loading control. n = 3. e. Infectivity assays were performed using cell lysates from Fig. 5a at different dilutions to infect fresh HeLa229 cells. 24 hpi the cells were fixed and the number of cells and number of inclusions were counted to plot the graph (box and whisker plots with a minimum and maximum range of values). n = 3. Paired t test (two tailed) was used for analysis p value <0.0001 indicates ****, t = 36,53, df=2. f. HeLa229 cells were either grown with or without glutamine/nucleosides (NS) as supplements. The cells were left uninfected or infected with Chlamydia at an MOI 1 for 30 h. The cells were lysed and analysed by western blotting. cHsp60 indicates the infection level. ß Actin serve as the loading control. n = 3.
Extended Data Fig. 6 Chlamydia metabolically reprograms the host cell.
a. HeLa229 cells were either left uninfected (UI) or infected with living (Ct) or heat inactivated Ct for 24 h. The cells were lysed and used for Western blot analysis to detect the levels of c-Myc and SLC1A5. cHsp60 indicates the infection level. ß actin serve as the loading control. n = 3. b. Infectivity assay from Fig. 6i. HeLa229 cells carrying an inducible shRNA expression cassette to silence c-Myc expression were either induced with AHT or left uninduced. These cells were in addition transfected with an expression plasmid for SLC1A5. The cells were infected with Chlamydia with an MOI 1 for 30 h. The number of inclusions was counted to plot the graph presented in box and whisker. p value was determined by one-way ANOVA with Tukey’s multiple comparisons test (F = 59.32, Df=8). Error bar was defined as mean with SEM. The cell lysate from the corresponding experiment was used to analyse cHsp60 levels via Western blotting. c. The organoids from BL6 (WT), SLC1A5 + / + or SLC1A5 -/- mice were used to analyse the amino acid content by LC-MS. The levels of amino acids in the cells were used to plot the graph, normalized to BL6 (WT) mice values. Two independent experiments where done, n = 2. Error bar was defined as mean with SEM. d. Progeny from infected mouse organoids: Organoids were derived from SLC1A5 + / + or SLC1A5 -/- mice and infected with Chlamydia for 6 days. Then, the organoids were lysed and released bacteria were used to infect freshly plated HeLa cells. The cells were fixed and stained for Chlamydia (Ct, green) and phalloidin (red) n = 3. Scale bar = 10 µm. Representative images of three independent experiments are shown.
Supplementary information
Supplementary Information
Supplementary Table 1 (containing primers for qPCR).
Source data
Source Data Fig. 1
Statistical source data for Fig. 1
Source Data Fig. 2
Statistical source data for Fig. 2
Source Data Fig. 3
Unmodified gels for Fig. 3
Source Data Fig. 4
Unmodified gels for Fig. 4
Source Data Fig. 5
Unmodified gels for Fig. 5
Source Data Fig. 6
Unmodified gels for Fig. 6
Source Data Extended Data Fig. 1
Statistical source data for Extended Data Fig. 1
Source Data Extended Data Fig. 3
Statistical source data for Extended Data Fig. 3
Source Data Extended Data Fig. 4
Statistical source data for Extended Data Fig. 4
Source Data Extended Data Fig. 5
Statistical source data for Extended Data Fig. 5
Source Data Extended Data Fig. 6
Statistical source data for Extended Data Fig. 6
Rights and permissions
About this article
Cite this article
Rajeeve, K., Vollmuth, N., Janaki-Raman, S. et al. Reprogramming of host glutamine metabolism during Chlamydia trachomatis infection and its key role in peptidoglycan synthesis. Nat Microbiol 5, 1390–1402 (2020). https://doi.org/10.1038/s41564-020-0762-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-0762-5
This article is cited by
-
Intracellular lifestyle of Chlamydia trachomatis and host–pathogen interactions
Nature Reviews Microbiology (2023)
-
Chlamydien als Risikofaktoren für Eierstock- und Gebärmutterhalskrebs
BIOspektrum (2023)
-
Organoids as host models for infection biology – a review of methods
Experimental & Molecular Medicine (2021)