Abstract
Porphyromonas gingivalis, an asaccharolytic member of the Bacteroidetes, is a keystone pathogen in human periodontitis that may also contribute to the development of other chronic inflammatory diseases. P. gingivalis utilizes protease-generated peptides derived from extracellular proteins for growth, but how these peptides enter the cell is not clear. Here, we identify RagAB as the outer-membrane importer for these peptides. X-ray crystal structures show that the transporter forms a dimeric RagA2B2 complex, with the RagB substrate-binding surface-anchored lipoprotein forming a closed lid on the RagA TonB-dependent transporter. Cryo-electron microscopy structures reveal the opening of the RagB lid and thus provide direct evidence for a ‘pedal bin’ mechanism of nutrient uptake. Together with mutagenesis, peptide-binding studies and RagAB peptidomics, our work identifies RagAB as a dynamic, selective outer-membrane oligopeptide-acquisition machine that is essential for the efficient utilization of proteinaceous nutrients by P. gingivalis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data supporting the findings of this study are available from the corresponding authors upon reasonable request. Coordinates and structure factors that support the findings of this study have been deposited in the Protein Data Bank with accession codes 6SLI (KRAB RagAB), 6SLJ (WT RagAB) and 6SLN (WT RagAB + P21). Electron microscopy structure coordinates have been deposited in the Electron Microscopy Data Bank with accession codes EMD-6SM3 (CC), EMD-6SMQ (OC) and EMD-6SML (OO). The raw cryo-EM movie-mode micrographs for the primary dataset containing the CC, OC and OO structures will be deposited in the Electron Microscopy Public Image Archive. Source data for Figs. 1a, 4a–d and Extended Data Figs. 2, 6a,b and 7 are included in this article and its Supplementary Information files.
References
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C. & Kent, R. L. Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144 (1998).
Eke, P. I. et al. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J. Periodontol. 86, 611–622 (2015).
Dominy, S. S. et al. Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 5, eaau3333 (2019).
Potempa, J., Mydel, P. & Koziel, J. The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat. Rev. Rheumatol. 13, 606–620 (2017).
Tonetti, M. S. et al. Treatment of periodontitis and endothelial function. N. Engl. J. Med. 356, 911–920 (2007).
Usher, A. K. & Stockley, R. A. The link between chronic periodontitis and COPD: a common role for the neutrophil? BMC Med. 11, 241 (2013).
Bui, F. Q. et al. Association between periodontal pathogens and systemic disease. Biomed J. 42, 27–35 (2019).
Mayrand, D. & Holt, S. C. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol. Rev. 52, 134–152 (1988).
Nemoto, T. K., Ohara-Nemoto, Y., Bezerra, G. A., Shimoyama, Y. & Kimura, S. A. Porphyromonas gingivalis periplasmic novel exopeptidase, acylpeptidyl oligopeptidase, releases N-acylated di- and tripeptides from oligopeptides. J. Biol. Chem. 291, 5913–5925 (2016).
Potempa, J., Banbula, A. & Travis, J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol. 2000 24, 153–192 (2000).
Grenier, D. et al. Role of gingipains in growth of Porphyromonas gingivalis in the presence of human serum albumin. Infect. Immun. 69, 5166–5172 (2001).
Nagano, K. et al. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J. Med. Microbiol. 56, 1536–1548 (2007).
Goulas, T. et al. Structure of RagB, a major immunodominant outer-membrane surface receptor antigen of Porphyromonas gingivalis. Mol. Oral Microbiol. 31, 472–485 (2016).
Bolam, D. N. & van den Berg, B. TonB-dependent transport by the gut microbiota: novel aspects of an old problem. Curr. Opin. Struct. Biol. 51, 35–43 (2018).
Bolam, D. N. & Koropatkin, N. M. Glycan recognition by the Bacteroidetes Sus-like systems. Curr. Opin. Struct. Biol. 22, 563–569 (2012).
Glenwright, A. J. et al. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541, 407–411 (2017).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Noinaj, N., Guillier, M., Barnard, T. J. & Buchanan, S. K. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 (2010).
Freed, D. M., Horanyi, P. S., Wiener, M. C. & Cafiso, D. S. Conformational exchange in a membrane transport protein is altered in protein crystals. Biophys. J. 99, 604–610 (2010).
Hickman, S. J., Cooper, R. E. M., Bellucci, L., Paci, E. & Brockwell, D. J. Gating of TonB-dependent transporters by substrate-specific forced remodelling. Nat. Commun. 8, 14804 (2017).
Gómez-Santos, N., Glatter, T., Koebnik, R., Swiątek-Połatyńska, M. A. & Søgaard-Andersen, L. A. TonB-dependent transporter is required for secretion of protease PopC across the bacterial outer membrane. Nat. Commun. 10, 1360 (2019).
Hall, L. M. et al. Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis. Infect. Immun. 73, 4253–4262 (2005).
Milner, P., Batten, J. E. & Curtis, M. A. Development of a simple chemically defined medium for Porphyromonas gingivalis: requirement for α-ketoglutarate. FEMS Microbiol. Lett 140, 125–130 (1996).
Grenier, D. et al. Role of gingipains in growth of Porphyromonas gingivalis in the presence of human serum albumin. Infect. Immun. 69, 5166–5172 (2001).
Nagano, K. et al. Trimeric structure of major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis. J. Bacteriol. 187, 902–911 (2005).
Lamster, I. B. & Ahio, J. K. Analysis of gingival crevicular fluid as applied to the diagnosis of oral and systemic diseases. Ann. NY Acad. Sci. 1098, 216–229 (2007).
Curtis, M. A., Hanley, S. A. & Aduse-Opoku, J. The rag locus of Porphyromonas gingivalis: a novel pathogenicity island. J. Periodontal Res. 34, 400–405 (1999).
Josts, I., Veith, K. & Tidow, H. Ternary structure of the outer membrane transporter FoxA with resolved signalling domain provides insights into TonB-mediated siderophore uptake. eLife 8, e48528 (2019).
Kim, I., Stiefel, A., Plantör, S., Angerer, A. & Braun, V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol. Microbiol. 23, 333–344 (1997).
Braun, V., Mahren, S. & Ogierman, M. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 6, 173–180 (2003).
Koebnik, R. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13, 343–347 (2005).
Ho, M. H., Lamont, R. J. & Xie, H. Identification of Streptococcus cristatus peptides that repress expression of virulence genes in Porphyromonas gingivalis. Sci. Rep. 7, 1413 (2017).
Moynié, L. et al. The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model. Nat. Commun. 10, 3673 (2019).
Balhesteros, H. et al. TonB-Dependent Heme/Hemoglobin Utilization by Caulobacter crescentus HutA. J. Bacteriol. 199, e00723–16 (2017).
Koropatkin, N. M., Martens, E. C., Gordon, J. I. & Smith, T. J. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–1115 (2008).
Rogowski, A. et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun. 6, 7481 (2015).
Nguyen, K. A., Travis, J. & Potempa, J. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-negative bacteria? J. Bacteriol. 189, 833–843 (2007).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Chiu, J., March, P. E., Lee, R. & Tillett, D. Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32, e174 (2004).
Tagawa, J. et al. Development of a novel plasmid vector pTIO-1 adapted for electrotransformation of Porphyromonas gingivalis. J. Microbiol. Methods 105, 174–179 (2014).
Belanger, M., Rodrigues, P. & Progulske-Fox, A. Genetic manipulation of Porphyromonas gingivalis. Curr. Protoc. Microbiol. 5, 13C.2.1–13C.2.24 (2007).
Smith, C. J. Genetic transformation of Bacteroides spp. using electroporation. Methods Mol. Biol 47, 161–169 (1995).
Filip, C., Fletcher, G., Wulff, J. L. & Earhart, C. F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115, 717–722 (1973).
Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995).
Winter, G., Lobley, C. M. & Prince, S. M. Decision making in xia2. Acta Crystallogr. D 69, 1260–1273 (2013).
Winter, G. et al. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr. D 74, 85–97 (2018).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Li, X., Zheng, S. Q., Egami, K., Agard, D. A. & Cheng, Y. Influence of electron dose rate on electron counting images recorded with the K2 camera. J. Struct. Biol. 184, 251–260 (2013).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Thompson, R. F., Iadanza, M. G., Hesketh, E. L., Rawson, S. & Ranson, N. A. Collection, pre-processing and on-the-fly analysis of data for high-resolution, single-particle cryo-electron microscopy. Nat. Protoc. 14, 100–118 (2019).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Petterson, E. F. et al. USCF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Darden, T., York, D. & Pedersen, L. Particle mesh ewald: an N log (N) method for ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Nosé, S. A. Molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 52, 255–268 (1984).
Hoover, W. G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A 31, 1695 (1985).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Tian, W., Chen, C., Lei, X., Zhao, J. & Liang, J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367 (2018).
Acknowledgements
This work was supported by a Wellcome Trust Investigator award (no. 214222/Z/18/Z to B.v.d.B.). We thank personnel of the Diamond Light Source for beam time (Block Allocation Group nos. mx-13587 and mx-18598) and assistance with data collection. All electron microscopy was performed at the Astbury Biostructure Laboratory which was funded by the University of Leeds and the Wellcome Trust (108466/Z/15/Z). We thank R. Thompson, E. Hesketh and D. Maskell for electron microscopy support and T. Kantyka for help in designing mass spectrometry experiments. This study was supported in part by National Science Centre, Poland grants UMO-2015/19/N/NZ1/00322 and UMO-2018/28/T/NZ1/00348 to M.M., UMO-2016/23/N/NZ1/01513 to Z.N., UMO-2018/29/N/NZ1/00992 to G.P.B., and UMO-2018/31/B/NZ1/03968 and NIDCR/DE 022597 (NIH) to J.P. LC–MS/MS analysis was supported by the Novo Nordisk Foundation (BioMS). J.B.R.W. was supported by a Wellcome Trust four-year PhD studentship (215064/Z/18/Z).
Author information
Authors and Affiliations
Contributions
M.M., J.P. and B.v.d.B. initiated the project. M.M. cultured cells, purified and crystallized proteins and performed MST binding experiments with guidance from J.P. and B.v.d.B. J.B.R.W. and S.R. determined cryo-EM structures, supervised by N.A.R. Z.N. performed cloning and strain construction. G.P.B. carried out quantitative PCR experiments, and C.S. and J.J.E. performed the peptidomics analysis. K.P. performed the molecular dynamics simulations, supervised by U.K. B.v.d.B. purified and crystallized proteins and determined the RagAB crystal structures. A.B. collected crystallography data. The manuscript was written by B.v.d.B. with input from M.M., J.B.R.W., N.A.R. and J.P.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
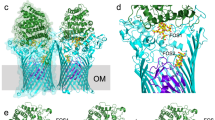
Extended Data Fig. 1 Unbiased peptide density in RagAB.
Stereo diagrams showing 2Fo-Fc (a) and Fo-Fc density (b) in RagAB KRAB before any modelling and refinement of the peptide. Selected segments of RagA (cyan) and RagB (green) neighbouring the peptide are shown. Map contouring parameters are 1.0σ, carve = 2 for the 2Fo-Fc and 3.0σ, carve = 2 for the Fo-Fc map. The extensive contacts of RagA with the peptide are confirmed by a PISA interface analysis17 which shows that 26 RagA residues form an interface with the peptide compared to only 8 for RagB, generating interface areas of 620 and 240 Å2 with RagA and RagB respectively. The PISA CSS (complexation significance score) is maximal (1.0) for peptide-RagA while it is only 0.014 for peptide-RagB. This suggests that the observed co-crystal structure represents a state where the ligand has been partially transferred from an initial, presumably low(er)-affinity binding site on RagB to a high(er)-affinity binding site in the RagAB complex, allowing co-purification.
Extended Data Fig. 2 RagAB binds a wide range of oligopeptides.
a-c, LC-MS/MS analysis of peptides bound to RagAB W83 KRAB, showing length distribution (a), total charge (b) and pI (c). d-f, Analysis of peptides bound to RagAB W83 wild-type, showing length distribution (d), total charge (e) and pI (f). For charge calculations, the pH was assumed to be 7.0 and contributions of any His residues were ignored. g, Amino acid frequency of RagAB-bound peptides (KRAB and wild-type combined; black) vs. the amino acid composition in the P. gingivalis proteome (gray), showing a substantial enrichment of Ala, Glu, Lys, Thr and Val. By contrast, aromatics (Phe, Trp) and bulky hydrophobics (Leu) appear to be under-represented. The peptides bound to RagAB from W83 KRAB vary in length from 7 to 29 residues, with a broad maximum of around 13 residues that fits well with the peptide density observed in the structures. Assuming equal abundance of each detected peptide, there is a slight preference for neutral to slightly acidic peptides, and the pI distribution has a bimodal shape, with maxima for acidic and slightly basic peptides. Analysis of the smaller RagAB-bound peptide set from wild-type W83 (d-f) yields a slightly wider size range from 5–36 residues, but overall there are no dramatic differences in the collective length, net charge and pI of the RagAB-bound peptide populations from W83 KRAB and wild-type strains.
Extended Data Fig. 3 Molecular dynamics simulations of RagAB show lid opening.
a, Cα-rmsd values of RagB lids in apo-RagAB (red) and peptide-bound RagAB (green) with reference to the starting crystal structure in the closed conformation. The Cα-rmsd values of the RagB lids in RagA2B2 are shown in blue with reference to the OO EM state. Each point indicates an average of 50 ns simulation trajectory. b, Comparison of the RagA2B2 open conformation from EM (magenta) with the snapshot of the most open simulation at 2500 ns (green). c, Internal surface of peptide binding cavities in closed holo-RagAB and apo-RagAB, generated with CASTp68. The bound peptide from a RagAB subunit in the crystal structure was removed in silico to generate a closed apo-complex, and three independent MD simulations were performed. For one of the simulations, a clear opening of the RagB lid was observed, reminiscent of recent results for a SusCD transporter and supporting the notion that ligand removal resets the transporter to favour the open state16. None of the peptide-bound complexes shows lid opening on the timescale of the simulations. While this suggests that lid opening is less favourable in the ligand-bound state, it does not contradict our observation of open, ligand-bound complexes via EM. The EM structures allowed us to compare both open states, which showed that the RagB lid in the simulation opens less wide than that in the EM structure, at least during the timescale of the simulation. We also observed a partial closing of both RagB lids during a 1000 ns simulation starting from the OO EM state (a, blue curves). The r.m.s.d. values of both RagB subunits decrease from ~30 Å in the EM structure (t = 0 ns) to ~15 Å, which is similar to the opening observed in one of the apo-RagAB simulations starting from the closed structure. Thus, it appears that the energy minimum for the open state in the simulations is different from that in solution, for reasons that are not clear.
Extended Data Fig. 4 RagB moves as a rigid body during lid opening.
a, Superposition of RagA subunits in the open (yellow) and closed (cyan) RagAB complexes in the OC state, showing the rigid-body movement of RagB. Equivalent points are indicated by x,y,z (closed RagB; green) and by x*, y*, z* (open RagB; red). The arrow indicates the approximate pivot point in the N-terminus of RagB at the back of the complex. b, Superposition as in (a), viewed from the extracellular side. Lid opening results in displacements of up to 45 Å for main chain atoms at the front of the complex, furthest away from the RagB N-terminus. c, Superposition of the open and closed states of RagB, with the N-termini indicated. d, Extracellular view of superposed RagA, with selected loops labelled. The conformational changes upon lid movement are mostly confined to those parts of the protein that continue to interact with RagB at the back of the complex (L7-L9).
Extended Data Fig. 5 Local resolution-filtered cryoEM maps and evidence for NTE density.
a, CC, OC and OO states of RagAB filtered and coloured by local resolution. Corresponding Fourier Shell Correlation (FSC) curves are shown (right). b, Unsharpened maps of RagAB displayed at low contour levels to reveal diffuse density attributed to the NTE. CC, OC and OO states are coloured purple, blue and green respectively.
Extended Data Fig. 6 Analysis of RagAB mutants.
a, Representative growth curves (n = 3, mean ± standard error of the mean) for mutant W83 ragAB variants on BSA-MM. For comparison, W83 WT and ΔragAB strains are shown as well. b, Representative SDS-PAGE gel (n = 2) showing OM protein expression levels following removal of inner membrane proteins by sarkosyl treatment. The RagABmono and both RagA hinge loop mutant strains have a similar phenotype as ΔRagAB, suggesting they cannot take up oligopeptides produced by gingipains. However, the OM protein levels show that very little RagAB is present, so that no conclusions about functionality can be drawn. The RagB acidic loop mutants show intermediate growth on BSA-MM, suggesting that oligopeptide uptake is somewhat impaired. However, the OMP levels of the acidic loop mutants are substantially lower than wild type, suggesting that both acidic loop mutants are likely functional and arguing that the slow growth of rag-4 P. gingivalis strain ATCC 33277 on BSA-MM is not due to the absence of the acidic loop in RagB.
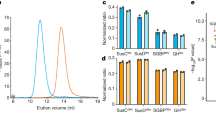
Extended Data Fig. 7 RagAB and RagB bind peptides selectively.
MST titration curves with the P4-FAM peptide for (a) RagAB from ATCC 33277, (b) RagAB from W83 and (c) Omp40-41 from W83 (negative control). The other panels show MST profiles for unlabelled P21, P12 or P4 binding to His-tag labelled W83 RagAB (d-f) and His-tag labelled W83 RagB (g-i). Experiments and listed Kd values represent the mean of three independent experiments ± SD.
Extended Data Fig. 8 Electron density comparison for different peptide ensembles.
a, Fo-Fc electron density maps (contoured at 3.0 σ) for the modelled peptide following final refinement for RagAB purified from W83 KRAB (left panel; 3.4 Å resolution), RagAB from W83 wild type co-crystallised with excess P21 peptide (middle panel; 2.6 Å), and RagAB from W83 wild type (right panel; 3.0 Å). Peptide sequence of the P21 co-crystal structure is arbitrarily modelled as QNGGANTSRGSAG, with numbering in italics according to the W83 KRAB peptide. Neighbouring residues D400-M407 of RagA are shown as cyan stick models for orientation purposes. b, Simulated annealing composite 2Fo-Fc omit maps for peptides bound to RagAB complexes as in (a). An annealing temperature of 500 K was used, with 5% of the models omitted. The fact that two different peptide ensembles produce similar maps as the P21 peptide, together with the inability to model the P21 sequence, suggests that the substrates are bound with register shifts and perhaps different chain directions. c, Stereo view of superposed 2Fo-Fc maps (made within Phenix; 1.0 σ, carve = 2.0) for WT RagAB in the absence (blue) and presence (orange) of P21 peptide, generated with the same high resolution cutoff (3.0 Å). For orientation purposes, the P21 peptide model is shown as sticks.
Extended Data Fig. 9 Schematic demonstrating the proposed mechanism of substrate capture and translocation by RagAB.
Peptide ligands to be imported by the RagAB system are predominantly generated by the action of gingipains on serum and tissue-derived proteins. 1. A lid-open state of RagAB permits peptide binding. 2. Contributions from both RagA and RagB to peptide binding elicits closure of the lid, forming the transport-competent state of the complex. This is signalled across the OM by perturbation of the TonB box region on the periplasmic side of the plug domain, making it accessible to TonB. 3. According to the literature consensus, TonB-mediated disruption of the plug permits substrate translocation and a return to the open state of RagAB.
Extended Data Fig. 10 Cryo-electron microscopy data support substrate-induced lid closure in RagAB.
a,b, 3D classes for RagAB ‘as purified’ (a) and in the presence of 50-fold excess P21 peptide (b). Classes corresponding to the CC, OC and OO states are coloured purple, blue and green respectively. Junk or ambiguous classes e.g. where RagA barrels are incomplete are coloured grey. In the presence of P21 there was no clear OO state whilst the the proportion of the CC state increased, supporting the proposed mechanism of substrate capture.
Supplementary information
Supplementary Information
Supplementary Figures 1–3, Tables 1–4 and Video legend.
Supplementary Video
Dynamics of the RagAB transporter. Cryo-EM map of open–closed (OC) RagAB, with cartoon models shown for RagA (blue) and RagB (yellow or grey) subunits.
Supplementary Data
Supplementary Table 2 RagAB peptidomics and in vitro binding of peptides to RagAB and RagB. Both analyses contain two separate Excel spreadsheets: all information retrieved from Mascot (Mascot) and a reduced spreadsheet with summed spectra (duplicates) and one charge variant of each peptide (Spectral C.). For spectral C., additional statistics were calculated: spectral count peptide, summed number of spectra of particular peptide; spectral count protein, summed number of spectra per particular protein; spectral count sample, summed number of spectra per particular sample; ratio peptide/protein, ratio of total number of particular peptide spectra to total number of spectra per protein; ratio peptide/sample, ratio of total number of particular peptide spectra to total number of spectra per protein.
Source data
Source Data Fig. 1
Unprocessed SDS–PAGE gel for Fig. 1a.
Source Data Fig. 4
Excel spreadsheet with source data for Fig. 4.
Source Data Extended Data Fig. 2
Excel spreadsheet with source data for Extended Data Fig. 2.
Source Data Extended Data Fig. 6
Excel spreadsheet with source data for Extended Data Fig. 6a.
Source Data Extended Data Fig. 6
Unprocessed SDS–PAGE gel for Extended Data Fig. 6b.
Source Data Extended Data Fig. 7
Excel spreadsheet with source data for Extended Data Fig. 7.
Rights and permissions
About this article
Cite this article
Madej, M., White, J.B.R., Nowakowska, Z. et al. Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis. Nat Microbiol 5, 1016–1025 (2020). https://doi.org/10.1038/s41564-020-0716-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-0716-y
This article is cited by
-
Identification and cultivation of anaerobic bacterial scavengers of dead cells
The ISME Journal (2023)
-
BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B12 uptake in gut Bacteroides
Nature Communications (2023)
-
Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes
Nature (2023)
-
Anti-biofilm effect of salivary histatin 5 on Porphyromonas gingivalis
Applied Microbiology and Biotechnology (2023)
-
Immunoelectron Microscopic Analysis of Dipeptidyl-Peptidases and Dipeptide Transporter Involved in Nutrient Acquisition in Porphyromonas gingivalis
Current Microbiology (2023)