Abstract
CRISPR–Cas adaptive immune systems protect bacteria and archaea against their invading genetic parasites, including bacteriophages/viruses and plasmids. In response to this immunity, many phages have anti-CRISPR (Acr) proteins that inhibit CRISPR–Cas targeting. To date, anti-CRISPR genes have primarily been discovered in phage or prophage genomes. Here, we uncovered acr loci on plasmids and other conjugative elements present in Firmicutes using the Listeria acrIIA1 gene as a marker. The four identified genes, found in Listeria, Enterococcus, Streptococcus and Staphylococcus genomes, can inhibit type II-A SpyCas9 or SauCas9, and are thus named acrIIA16–19. In Enterococcus faecalis, conjugation of a Cas9-targeted plasmid was enhanced by anti-CRISPRs derived from Enterococcus conjugative elements, highlighting a role for Acrs in the dissemination of plasmids. Reciprocal co-immunoprecipitation showed that each Acr protein interacts with Cas9, and Cas9–Acr complexes were unable to cleave DNA. Northern blotting suggests that these anti-CRISPRs manipulate single guide RNA length, loading or stability. Mirroring their activity in bacteria, AcrIIA16 and AcrIIA17 provide robust and highly potent broad-spectrum inhibition of distinct Cas9 proteins in human cells (for example, SpyCas9, SauCas9, SthCas9, NmeCas9 and CjeCas9). This work presents a focused analysis of non-phage Acr proteins, demonstrating a role in horizontal gene transfer bolstered by broad-spectrum CRISPR–Cas9 inhibition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article and its Extended Data, Source Data and Supplementary Information files. All relevant accession codes are available in Table 1.
Change history
30 April 2020
In the version of this Article originally published, Extended Data Fig. 4 was missing a label for panel c; this has now been corrected.
23 April 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41564-020-0726-9
References
Palmer, K. L., Kos, V. N. & Gilmore, M. S. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 13, 632–639 (2010).
Waldor, M. K. & Mekalanos, J. J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914 (1996).
Koonin, E. V. Viruses and mobile elements as drivers of evolutionary transitions. Philos. Trans. R. Soc. Lond. B 371, 20150442 (2016).
Labrie, S. J., Samson, J. E. & Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 (2010).
PriceV. J., Huo W., Sharifi A. & Palmer K. L. CRISPR-cas and restriction-modification act additively against conjugative antibiotic resistance plasmid transfer in enterococcus faecalis. mSphere 1, e00064-16 (2016).
Edgar, R. & Qimron, U. The Escherichia coli CRISPR system protects from λ lysogenization, lysogens, and prophage induction. J. Bacteriol. 192, 6291–6294 (2010).
Zhang, Y. et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol. Cell 50, 488–503 (2013).
Makarova, K. S. et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13, 722–736 (2015).
Garneau, J. E. et al. The CRISPR/cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010).
Clark, D. P. & Pazdernik, N. J. in Molecular Biology 2nd edn, Ch. e26 (Elsevier, 2013).
Casjens, S. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49, 277–300 (2003).
Bondy-Denomy, J., Pawluk, A., Maxwell, K. L. & Davidson, A. R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013).
Zhu, Y. et al. Diverse mechanisms of CRISPR-Cas9 inhibition by type IIC Anti-CRISPR proteins. Mol. Cell 74, 296–309 (2019).
Thavalingam, A. et al. Inhibition of CRISPR–Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2. Nat. Commun. 10, 2806 (2019).
Bondy-Denomy, J. et al. Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139 (2015).
Harrington, L. B. et al. A broad-spectrum inhibitor of CRISPR-Cas9. Cell 170, 1224–1233 (2017).
Dong, L. et al. An anti-CRISPR protein disables type V Cas12a by acetylation. Nat. Struct. Mol. Biol. 26, 308–314 (2019).
Knott, G. J. et al. Broad-spectrum enzymatic inhibition of CRISPR–Cas12a. Nat. Struct. Mol. Biol. 26, 315–321 (2019).
Rauch, B. J. et al. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 168, 150–158 (2017).
Hynes, A. P. et al. An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nat. Microbiol. 2, 1374–1380 (2017).
Hynes, A. P. et al. Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat. Commun. 9, 2919 (2018).
Uribe, R. V. et al. Discovery and characterization of Cas9 inhibitors disseminated across seven bacterial phyla. Cell Host Microbe 25, 233–241 (2019).
Forsberg, K. J. et al. Functional metagenomics-guided discovery of potent cas9 inhibitors in the human microbiome. eLife 8, e46540 (2019).
Pawluk, A. et al. Naturally occurring off-switches for CRISPR-Cas9. Cell 167, 1829–1838 (2016).
Jiang, F. et al. Temperature-responsive competitive inhibition of CRISPR-Cas9. Mol. Cell 73, 601–610 (2019).
Liu, L., Yin, M., Wang, M. & Wang, Y. Phage AcrIIA2 DNA mimicry: structural basis of the CRISPR and anti-CRISPR arms race. Mol. Cell 73, 611–620 (2019).
Dong, D. et al. Structural basis of CRISPR–SpyCas9 inhibition by an anti-CRISPR protein. Nature 546, 436–439 (2017).
Shin, J. et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 3, e1701620 (2017).
Garcia, B. et al. Anti-CRISPR AcrIIA5 potently inhibits all Cas9 homologs used for genome editing. Cell Rep. 29, 1739–1746 (2019).
Borges, A. L. et al. Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 Immunity. Cell 174, 917–925 (2018).
Osuna, B. A. et al. Listeria phages induce Cas9 degradation to protect lysogenic genomes. Preprint at https://doi.org/10.1101/787200 (2019).
Hullahalli, K., Rodrigues, M. & Palmer, K. L. Exploiting CRISPR-Cas to manipulate Enterococcus faecalis populations. eLife 6, e26664 (2017).
Trasanidou, D. et al. Keeping crispr in check: diverse mechanisms of phage-encoded anti-crisprs. FEMS Microbiol. Lett. 366, 1–14 (2019).
Zhang, F., Song, G. & Tian, Y. Anti-CRISPRs: the natural inhibitors for CRISPR-Cas systems. Anim. Model. Exp. Med. 2, 69–75 (2019).
PalmerK. L. & GilmoreM. S. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1, e00227-10 (2010).
Hullahalli, K., Rodrigues, M., Nguyen, U. T. & Palmer, K. An attenuated CRISPR-cas system in enterococcus faecalis permits DNA acquisition. mBio 9, e00414-18 (2018).
Seamon, K. J., Light, Y. K., Saada, E. A., Schoeniger, J. S. & Harmon, B. Versatile high-throughput fluorescence assay for monitoring Cas9 activity. Anal. Chem. 90, 6913–6921 (2018).
Hoang, T. T., Kutchma, A. J., Becher, A. & Schweizer, H. P. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72 (2000).
Choi, K. H., Kumar, A. & Schweizer, H. P. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397 (2006).
Bhardwaj, P., Ziegler, E. & Palmer, K. L. Chlorhexidine induces VanA-type Vancomycin resistance genes in enterococci. Antimicrob. Agents Chemother. 60, 2209–2221 (2016).
Kleinstiver, B. P. et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282 (2019).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Acknowledgements
We thank K. L. Palmer (UT Dallas) and G. M. Dunny (University of Minnesota) for providing all E. faecalis strains, vector and protocols. We thank M. Johnson for his assistance in executing PlasFlow, N. Marino for her profound wisdom and comedic wit, and all members of the Bondy-Denomy Lab for their continuous support and thoughtful discussions contributing to this study. Bondy-Denomy Lab was supported by the UCSF Program for Breakthrough Biomedical Research funded in part by the Sandler Foundation, the Searle Fellowship, the Vallee Foundation, an NIH Director’s Early Independence Award No. DP5-OD021344, and NIH grant no. R01GM127489. Research in the Kleinstiver Lab is supported by NIH grant no. R00-CA218870, an A.S.G.C.T. Career Development Award and the Margaret Q. Landenberger Research Foundation. This research was carried out using Defense Advanced Research Projects Agency (DARPA) award HR0011-17-2-0043. The views, opinions and/or findings should be attributed to the author and should not be interpreted as representing the official views or policies of the Department of Defense or the US Government.
Author information
Authors and Affiliations
Contributions
C.M. and J.B.-D. conceived and designed the study. C.M. conducted bioinformatic, bacterial and biochemical experiments. K.A.C. performed human cell experiments. B.A.O. and R.P.-R. assisted with experimental design. B.P.K and J.B.-D. supervised experiments. C.M. and J.B.-D. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics, and a scientific advisory board member and co-founder of Acrigen Biosciences. B.P.K. is an inventor on various patents and patent applications that describe gene editing and epigenetic editing technologies, is a consultant for Avectas Inc. and an advisor for Acrigen Biosciences.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Schematic of acr loci and lethal self-genome cleavage assay.
a, Full schematic of acr loci with relevant neighboring genes displayed. b, Schematic of SpyCas9 in P. aeruginosa programmed to cause lethal self-genome cleavage to assess bacterial survival in the presence of AcrIIA proteins. CRISPR strength is determined by titrating levels of IPTG, which induces expression of sgRNA targeting the chromosomal phzM gene from a multicopy plasmid. OD600 measurements are represented as the mean of three biological replicates ± SD.
Extended Data Fig. 2 Anti-CRISPR distribution in integrative mobile genetic elements across bacterial taxa.
Phylogenetic analysis based on acrIIA16–19 homologs (panels a to d, respectively) reconstructed from a midpoint rooted minimum-evolution of full length protein sequences identified following an iterative PSI-BLASTp search, see methods for details. Number of genomes included to construct each tree for acrIIA16–19 are seventy, twenty-six, eighty-four and seventeen respectively. Branches are labeled with species name and colored according to species class (see legend). Species for which AcrIIA homologs have been tested in this study are shown in bold.
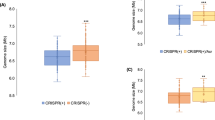
Extended Data Fig. 3 AcrIIA enhance conjugation-mediated horizontal gene transfer in E. faecalis, related to Fig. 2.
a, b. Mating outcomes during plasmid conjugation of a targeted plasmid from donor to recipient cells where indicated acrIIA genes are (a) pre-expressed in recipient cells, or (b) encoded on conjugating plasmid. Data displayed as 10-fold colony serial dilution spots of donor, recipient or transconjugant cells on selective antibiotic plates. Mating assays were performed in biological triplicate and produced similar outcomes. c, Schematic of E. faecalis conjugation of protospacer and acrIIA-bearing plasmid transferring into CRISPR-defective recipients. For CRISPR1, the bona fide AcrIIA4 is utilized to suppress CRISPR targeting, and a ΔCas9 strain from previously reported work is used for CRISPR3 (Price et al.,5). Red * denotes plasmids that have lost conjugation ability. Mating assays were performed in biological duplicate or triplicate and produced similar outcomes.
Extended Data Fig. 4 AcrIIA16–19 biochemical analysis, related to Figs. 3 and 4.
a, Coomassie-stained polyacrylamide gel showing GST-tagged AcrIIA proteins (IIA4 37kD, IIA16Lmo 50kD, IIA17Sga 39kD, IIA18Sma 48kD and IIA19Ssim 42kD) purified from E. coli by elution from Glutathione Sepharose columns. Visible bands at different sizes are co-purifying proteins from E. coli. Data shown are representative of two independent experiments. b, Coomassie-stained polyacrylamide gel showing co-immunoprecipitation of Acr proteins with Myc-tagged sgRNA-bound SpyCas9 pulled down from P. aeruginosa. Data shown are representative of two independent experiments. c, d, Uncropped versions of both Myc and GST pulldowns from Fig. 4a and c, displaying all fragments of (c) sgRNA-bound SpyCas9, or (d) Apo- SpyCas9 without sgRNA present. Data shown are representative of two independent experiments.
Extended Data Fig. 5 Acr inhibition activity in human cells tested against different Cas9 orthologs, related to Fig. 5.
Reported Acr proteins in this study and from previous works tested for inhibition of genome editing activities of Sth1Cas9, Sth3Cas9, Nme2Cas9 and CjeCas9 (a-d, respectively). Editing efficiencies against endogenous genes in HEK 293 T cells were assessed by targeted sequencing and quantified as the percentage of reads containing a nuclease-induced alteration; the no-Acr condition contains an EGFP expression plasmid; the NT control includes an empty U6 expression plasmid. Percent reads modified are represented as the mean of three biological replicates ± SD.
Supplementary information
Supplementary Information
Supplementary Information for Extended Data 2 phylogenetic tree analyses. Raw versions of the trees in NEXUS format.
Supplementary Tables
Supplementary Tables 1–4.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed gels and northern blots.
Source Data Fig. 4
Unprocessed gels and western blots.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed gels and western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Rights and permissions
About this article
Cite this article
Mahendra, C., Christie, K.A., Osuna, B.A. et al. Broad-spectrum anti-CRISPR proteins facilitate horizontal gene transfer. Nat Microbiol 5, 620–629 (2020). https://doi.org/10.1038/s41564-020-0692-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-0692-2
This article is cited by
-
Inhibitors of bacterial immune systems: discovery, mechanisms and applications
Nature Reviews Genetics (2024)
-
Bacteriophages suppress CRISPR–Cas immunity using RNA-based anti-CRISPRs
Nature (2023)
-
Optimization of Cas9 activity through the addition of cytosine extensions to single-guide RNAs
Nature Biomedical Engineering (2023)
-
Genome plasticity as a paradigm of antibiotic resistance spread in ESKAPE pathogens
Environmental Science and Pollution Research (2022)
-
Controlling and enhancing CRISPR systems
Nature Chemical Biology (2021)