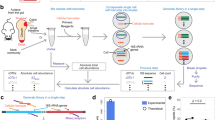
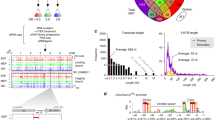
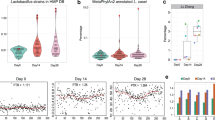
Abstract
A complex microbiota inhabits various microenvironments of the gut, with some symbiotic bacteria having evolved traits to invade the epithelial mucus layer and reside deep within the intestinal tissue of animals. Whether these distinct bacterial communities across gut biogeographies exhibit divergent behaviours is largely unknown. Global transcriptomic analysis to investigate microbial physiology in specific mucosal niches has been hampered technically by an overabundance of host RNA. Here, we employed hybrid selection RNA sequencing (hsRNA-Seq) to enable detailed spatial transcriptomic profiling of a prominent human commensal as it colonizes the colonic lumen, mucus or epithelial tissue of mice. Compared to conventional RNA-Seq, hsRNA-Seq increased reads mapping to the Bacteroides fragilis genome by 48- and 154-fold in mucus and tissue, respectively, allowing for high-fidelity comparisons across biogeographic sites. Near the epithelium, B. fragilis upregulated numerous genes involved in protein synthesis, indicating that bacteria inhabiting the mucosal niche are metabolically active. Further, a specific sulfatase (BF3086) and glycosyl hydrolase (BF3134) were highly induced in mucus and tissue compared to bacteria in the lumen. In-frame deletion of these genes impaired in vitro growth on mucus as a carbon source, as well as mucosal colonization of mice. Mutants in either B. fragilis gene displayed a fitness defect in competing for colonization against bacterial challenge, revealing the importance of site-specific gene expression for robust host-microbial symbiosis. As a versatile tool, hsRNA-Seq can be deployed to explore the in vivo spatial physiology of numerous bacterial pathogens or commensals.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
RNA-Seq and hsRNA-Seq data have been deposited in the NCBI SRA under accession no. PRJNA438372. The B. fragilis NCTC 9343 genome used for mapping is available at GenBank under assembly no. GCA_000025985.1. Source data for Figs. 1–4 and Source Data Extended Data Figs. 1, 8, 9 and 10 are included in this article.
Code availability
The code used in the analysis is available at https://github.com/wenchichou/bugInHost.
References
Topping, D. L. & Clifton, P. M. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81, 1031–1064 (2001).
Buffie, C. G. & Pamer, E. G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801 (2013).
Donaldson, G. P., Lee, S. M. & Mazmanian, S. K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 (2016).
Johansson, M. E. V. & Hansson, G. C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 16, 639–649 (2016).
Li, H. et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 6, 8292 (2015).
Wang, Y. et al. Regional mucosa-associated microbiota determine physiological expression of TLR2 and TLR4 in murine colon. PLoS ONE 5, e13607 (2010).
Yasuda, K. et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 17, 385–391 (2015).
Albenberg, L. et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147, 1055–1063.e8 (2014).
Davis, C. P. & Savage, D. C. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect. Immun. 10, 948–956 (1974).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Vélez, M. P., De Keersmaecker, S. C. J. & Vanderleyden, J. Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol. Lett. 276, 140–148 (2007).
Savage, D. C. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31, 107–133 (1977).
Rowan, F. et al. Bacterial colonization of colonic crypt mucous gel and disease activity in ulcerative colitis. Ann. Surg. 252, 869–875 (2010).
Pédron, T. et al. A crypt-specific core microbiota resides in the mouse colon. mBio 3, e00116-12 (2012).
Earle, K. A. et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18, 478–488 (2015).
Mark Welch, J. L., Hasegawa, Y., McNulty, N. P., Gordon, J. I. & Borisy, G. G. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl Acad. Sci. USA 114, E9105–E9114 (2017).
Marsh, J. W., Humphrys, M. S. & Myers, G. S. A. A laboratory methodology for dual RNA-sequencing of bacteria and their host cells in vitro. Front. Microbiol. 8, 1830 (2017).
Westermann, A. J., Barquist, L. & Vogel, J. Resolving host–pathogen interactions by dual RNA-seq. PLoS Pathog. 13, e1006033 (2017).
Humphrys, M. S. et al. Simultaneous transcriptional profiling of bacteria and their host cells. PLoS ONE 8, e80597 (2013).
Mavromatis, C. H. et al. The co-transcriptome of uropathogenic Escherichia coli-infected mouse macrophages reveals new insights into host–pathogen interactions. Cell. Microbiol. 17, 730–746 (2015).
Westermann, A. J. & Vogel, J. in Bacterial Regulatory RNA: Methods and Protocols (eds Arluison, V. & Valverde, C.) 59–75 (Springer, 2018).
Vannucci, F. A., Foster, D. N. & Gebhart, C. J. Laser microdissection coupled with RNA-seq analysis of porcine enterocytes infected with an obligate intracellular pathogen (Lawsonia intracellularis). BMC Genomics 14, 421 (2013).
Bernstein, J. A., Khodursky, A. B., Lin, P.-H., Lin-Chao, S. & Cohen, S. N. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl Acad. Sci. USA 99, 9697–9702 (2002).
Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–189 (2009).
Melnikov, A. et al. Hybrid selection for sequencing pathogen genomes from clinical samples. Genome Biol. 12, R73 (2011).
Matranga, C. B. et al. Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol. 15, 519 (2014).
Huang, J. Y., Lee, S. M. & Mazmanian, S. K. The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe 17, 137–141 (2011).
Lee, S. M. et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429 (2013).
Donaldson, G. P. et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800 (2018).
Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 (2016).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 (2017).
Blow, M. J. et al. The epigenomic landscape of prokaryotes. PLoS Genet. 12, e1005854 (2016).
Burroughs, A. M., Kaur, G., Zhang, D. & Aravind, L. Novel clades of the HU/IHF superfamily point to unexpected roles in the eukaryotic centrosome, chromosome partitioning, and biologic conflicts. Cell Cycle 16, 1093–1103 (2017).
Liu, C. H., Lee, S. M., Vanlare, J. M., Kasper, D. L. & Mazmanian, S. K. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc. Natl Acad. Sci. USA 105, 3951–3956 (2008).
Coyne, M. J., Chatzidaki-Livanis, M., Paoletti, L. C. & Comstock, L. E. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc. Natl Acad. Sci. USA 105, 13099–13104 (2008).
Round, J. L. et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 (2011).
Baughn, A. D. & Malamy, M. H. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427, 441–444 (2004).
Rocha, E. R. & Smith, C. J. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 181, 5701–5710 (1999).
Sund, C. J. et al. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol. Microbiol. 67, 129–142 (2008).
Schofield, W. B., Zimmermann-Kogadeeva, M., Zimmermann, M., Barry, N. A. & Goodman, A. L. The stringent response determines the ability of a commensal bacterium to survive starvation and to persist in the gut. Cell Host Microbe 24, 120–132.e6 (2011).
Benjdia, A., Martens, E. C., Gordon, J. I. & Berteau, O. Sulfatases and a radical S-adenosyl-l-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J. Biol. Chem. 286, 25973–25982 (2011).
Thomsson, K. A. et al. Detailed O-glycomics of the Muc2 mucin from colon of wild-type, core 1- and core 3-transferase-deficient mice highlights differences compared with human MUC2. Glycobiology 22, 1128–1139 (2012).
Scholz, M. et al. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat. Methods 13, 435–438 (2016).
Yassour, M. et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8, 343ra81 (2016).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Chu, H. et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120 (2016).
Round, J. L. & Mazmanian, S. K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 107, 12204–12209 (2010).
Jakobsson, H. E. et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 16, 164–177 (2015).
Thänert, R., Goldmann, O., Beineke, A. & Medina, E. Host-inherent variability influences the transcriptional response of Staphylococcus aureus during in vivo infection. Nat. Commun. 8, 14268 (2017).
Nuss, A. M. et al. Tissue dual RNA-seq allows fast discovery of infection-specific functions and riboregulators shaping host–pathogen transcriptomes. Proc. Natl Acad. Sci. USA 114, E791–E800 (2017).
Stapels, D. A. C. et al. Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 362, 1156–1160 (2018).
Aprianto, R., Slager, J., Holsappel, S. & Veening, J.-W. Time-resolved dual RNA-seq reveals extensive rewiring of lung epithelial and pneumococcal transcriptomes during early infection. Genome Biol. 17, 198 (2016).
Metsky, H. C. et al. Capturing sequence diversity in metagenomes with comprehensive and scalable probe design. Nat. Biotechnol. 37, 160–168 (2019).
Sonnenburg, J. L. et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 (2005).
Cao, Y., Rocha, E. R. & Smith, C. J. Efficient utilization of complex N-linked glycans is a selective advantage for Bacteroides fragilis in extraintestinal infections. Proc. Natl Acad. Sci. USA 111, 12901–12906 (2014).
Martens, E. C., Chiang, H. C. & Gordon, J. I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 (2008).
Kashyap, P. C. et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc. Natl Acad. Sci. USA 110, 17059–17064 (2013).
Pudlo, N. A. et al. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio 6, e01282-15 (2015).
Varel, V. H. & Bryant, M. P. Nutritional features of Bacteroides fragilis subsp. fragilis. Appl. Microbiol. 28, 251–257 (1974).
Kotarski, S. F. & Salyers, A. A. Isolation and characterization of outer membranes of Bacteroides thetaiotaomicron grown on different carbohydrates. J. Bacteriol. 158, 102–109 (1984).
Alexeyev, M. F. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26, 824–828 (1999).
Shishkin, A. A. et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods 12, 323–325 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338 (2017).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Lebreton, F. et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4, e00534-13 (2013).
Sefik, E. et al. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015).
Frith, M. C., Saunders, N. F. W., Kobe, B. & Bailey, T. L. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 4, e1000071 (2008).
Bailey, T. L. et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Lebreton, F. et al. Tracing the enterococci from paleozoic origins to the hospital. Cell 169, 849–861 (2017).
Chan, J. Z.-M., Halachev, M. R., Loman, N. J., Constantinidou, C. & Pallen, M. J. Defining bacterial species in the genomic era: insights from the genus Acinetobacter. BMC Microbiol. 12, 302 (2012).
Acknowledgements
We thank E. Hsiao, E. Martens, D. Gevers, C. Desjardins, B. Haas and J. Livny for helpful discussions, and members of the Mazmanian laboratory for comments. G.P.D. was supported by an NIH training grant no. 5T32 GM07616, National Science Foundation Graduate Research Fellowship no. DGE-1144469 and the Center for Environmental Microbial Interactions at Caltech. The project was funded by NIH grant no. U19AI110818 to the Broad Institute; NIH grant no. DK110534 to H.C.; NIH grant nos. GM099535 and DK078938, and the Heritage Medical Research Institute to S.K.M.
Author information
Authors and Affiliations
Contributions
G.P.D. and S.K.M. conceived the study. G.P.D., W.-C.C., G.G., A.M.E. and S.K.M. designed the study. G.P.D. prepared the samples for sequencing and performed the mouse colonization and microbiology experiments. D.C., P.R., J.B., A.M. and G.G. performed the hybrid capture and sequencing experiments. W.-C.C., A.L.M. and T.A. performed the computational analysis. H.C. performed the colitis model and flow cytometry. P.B.E. scored the sections for histology. G.P.D., W.-C.C. and A.L.M. created the figures. A.M.E. and S.K.M. supervised the work. G.P.D., W.-C.C., A.L.M., A.M.E. and S.K.M. wrote the paper. All authors provided input on the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Intestinal biogeography of Bacteroides fragilis during mono-colonization.
a, CFU per gram of lumen content and b, CFU per cm of mucus from indicated regions of intestine after 4 weeks of mono-colonization with wild-type B. fragilis (mean and standard error, n = 4 animals). c, CFU per sample in lumen, mucus, and tissue samples of the proximal intestine of mice mono-colonized for 4 weeks with wild-type B. fragilis (mean and standard error, n = 4 animals). These samples were collected using the same dissection method used to prepare samples for RNA-Seq (Fig. 1a).
Extended Data Fig. 2 Individual mouse correlation plots to assess hybrid selection performance.
Correlation plots for HS vs non-HS in individual mice (3 individual-mouse samples from lumen, 3 from mucus, and 3 from tissue, Pearson’s r). Each dot represents a single gene.
Extended Data Fig. 3 Host gene expression comparisons between samples with and without hybrid selection.
Total RNA-Seq reads were mapped to mm10 mouse genome using STAR, and the mapped reads were converted into read counts for each gene by HTSeq. After excluding genes with < 10 reads mapping across any sample, the read counts for each sample were normalized by TPM (Transcripts Per Million). Each dot represents a single gene. The average TPM for each gene is shown from non-hybrid selected libraries (x-axis) and hybrid selected libraries (y-axis) (n = 3 animals, Pearson’s r).
Extended Data Fig. 4 Normalized gene expression levels with and without hybrid selection are highly correlated with few outliers.
Each gene is represented by a single dot. The correlation coefficients for lumen, mucus, and tissue are 0.99, 0.96, and 0.98, respectively. Outliers where the difference between the HS and non-HS values is larger than three standard deviations are numbered and listed in Supplementary Table 3. These represent primarily short genes (median length 110 nucleotides), particularly tRNA and 5 s rRNA genes. Short genes (<200 nt) are colored blue, showing that most protein-coding genes are enriched properly.
Extended Data Fig. 5 Correlation in gene expression between different sample sites was improved with hybrid selection.
Each dot represents a single gene with all genes plotted (n = 3 animals, Pearson’s r).
Extended Data Fig. 6 Structural modeling for genes of interest using Phyre.
a, The predicted structure for BF3134, modeled using Phyre72, indicated that BF3134 is a likely cyclo-malto-dextrinase, closely related to neopullulanase and maltogenic amylase and a member of glycosyl hydrolase family 13 (96% of the sequence was modeled with 100% confidence to the cyclo-malto-dextrinase template c3edeB, with 42% identity). b, Secondary structure prediction for BF3134 using Phyre. Pfam domain analysis for BF3134 also indicated the presence of an N-terminal cyclo-malto-dextrinase domain (PF09087), a central alpha-amylase domain (glycosyl hydrolase family 13; PF00128), and a C-terminal cyclo-malto-dextrinase domain (PF10438). c, The predicted structure for BF3086 indicated a role as an acetylglucosamine-6-sulfatase (93% of the sequence was modeled with 100% confidence by the single highest scoring template, c5g2va, an n-acetylglucosamine-6-sulfatase, with 51% identity). d, Secondary structure prediction for BF3086. Pfam domain analysis indicated the presence of a sulfatase domain, in addition to a domain of unknown function (DUF4976) downstream of the sulfatase domain. The region aligned by Phyre with the c5g2va template included both the regions encompassed by the Pfam sulfatase domain, as well as the Pfam domain of unknown function (DUF4976).
Extended Data Fig. 7 BF3086 and BF3134 are conserved and share a potential regulatory motif.
a, Phylogeny of 92 Bacteroides and Parabacteroides strains74 showing the presence of BF3086 and BF3134 orthologues, with horizontal bar graphs indicating the percent protein sequence identity to the studied type strain (NCTC9343, highlighted with red font). The teal box indicates strains that can be confidently assigned to the B. fragilis species (average pairwise ANI78 between them is 98%, whereas it falls below 95% for the next-closest strains also labeled as B. fragilis). The black squares indicate the presence of the conserved upstream motif (0-2 mismatches), using the GLAM2Scan algorithm75. b, Sequence of the conserved motif upstream of both genes. The asterisk (*) at position 18 indicates a position that differs between the upstream regions of the glycosyl hydrolase (BF3086) and the sulfatase (BF3134). The glycosyl hydrolase upstream region has an “A” at this position, whereas the sulfatase upstream region has a deletion at this position.
Extended Data Fig. 8 Additional in vitro and in vivo phenotypes of ∆BF3086 and ∆BF3134.
a, BF3086 and BF3134 biological replicates. Fold-change for individual mice indicate consistently induced expression of BF3086 and BF3134 in the mucus and tissue relative to the lumen. b-e, Growth of individual B. fragilis strains in a defined minimal medium with b, inulin, c, pullulan, d, mannan, or e, pig mucin (mean and standard error, n = 8 independent cultures). f-h, Quantitative RT–PCR (∆∆Ct method normalized to gyrB) on fecal samples of mice mono-colonized with indicated strains of B. fragilis, assessing the expression of f, ccfC (BF3581), g, PSB flippase (BF1900), and h, PSC flippase (BF1014) (mean and standard error, Tukey ANOVA, n = 4 animals).
Extended Data Fig. 9 BF3134 is required for B. fragilis protection from experimental colitis.
a, Mice were mono-colonized with B. fragilis strains at weaning (3 weeks of age) before inducing DNBS colitis at 7 weeks of age. Body weights of mice were measured every 24 hours and are represented as a percentage of their starting weight on day 0 (Tukey 2-way ANOVA, n = 10, 9, 9, representative of two independent experiments). b, 72 hours after colitis induction, mice were sacrificed and the length of the colon from rectum to the cecal junction was dissected (representative images of 3 colons per group, images normalized to size using rulers and then cropped around the colon) and c, colon length measured (Tukey ANOVA, n = 10, 9, 9). d, Histopathologic scores of whole colons (max 48, mean and interquartile range, Tukey ANOVA, n = 10, 9, 9). e, Quantitative RT–qPCR (∆∆Ct method normalized to gyrB) on fecal samples of mice mono-colonized with indicated strains of B. fragilis, assessing the expression of the PSA flippase (BF1369) (Tukey ANOVA, n = 4 animals). f, Lymphocytes isolated from mesenteric lymph nodes of mono-colonized, DNBS-induced mice were analyzed using flow cytometry. IL-17A-producing T cells quantified as a percent of total CD4 + Foxp3 + regulatory T cells (Tukey ANOVA, n = 10, 9, 9 animals). g, IL-10-producing T cells quantified as a percent of total CD4 + Foxp3 + regulatory T cells (Tukey ANOVA, n = 10, 9, 9 animals, representative of two independent experiments) (all panels unless noted: mean and standard error, * p < 0.05, ** p < 0.01, *** p < 0.001).
Extended Data Fig. 10 Control experiments and flow cytometry methods for DNBS colitis.
a, Quantitative RT–qPCR (∆∆Ct method normalized to gyrB) for PSA flippase (BF1369) in lumen, mucus and tissue samples (mean and standard error, n = 4 animals). Fold-change between sample sites was quantified within each mouse individually. b, Mice mono-colonized with indicated strains of B. fragilis for one month were treated with 50% ethanol, the vehicle control for DNBS colitis induction. Mice were weighed every 24 hours, graphed as a percentage of their weight at day 0 (Tukey 2-way ANOVA, n = 5, 4, 4). c, 72 hours after treatment the mice were sacrificed and the length of the colon was measured from rectum to the cecal junction (Tukey 2-way ANOVA, n = 5, 4, 4) d, Example live cell gating for flow cytometry in Extended Data 9f and 9 g (representative from two independent experiments with similar results). e, Example flow plots (1 from each group) for assessing the proportion of IL-10 and IL-17 positive regulatory T cells, as quantified in Extended Data 9f and g (representative from two independent experiments with similar results, mean and standard error in graphs, * p < 0.05).
Supplementary information
Supplementary Information
Supplementary Tables 1, 2, 7, 9 and references. Descriptions for Supplementary Tables 3–6, 8 and 10.
Supplementary Table
Supplementary Tables 3–6, 8 and 10.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Donaldson, G.P., Chou, WC., Manson, A.L. et al. Spatially distinct physiology of Bacteroides fragilis within the proximal colon of gnotobiotic mice. Nat Microbiol 5, 746–756 (2020). https://doi.org/10.1038/s41564-020-0683-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-0683-3
This article is cited by
-
An expanded transcriptome atlas for Bacteroides thetaiotaomicron reveals a small RNA that modulates tetracycline sensitivity
Nature Microbiology (2024)
-
The gut microbiota and its biogeography
Nature Reviews Microbiology (2024)
-
Bacteroides fragilis toxin expression enables lamina propria niche acquisition in the developing mouse gut
Nature Microbiology (2024)
-
Maturation state of colonization sites promotes symbiotic resiliency in the Euprymna scolopes-Vibrio fischeri partnership
Microbiome (2023)
-
Interrogation of the mammalian gut–brain axis using LC–MS/MS-based targeted metabolomics with in vitro bacterial and organoid cultures and in vivo gnotobiotic mouse models
Nature Protocols (2023)