Abstract
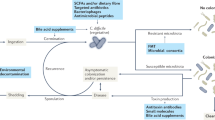
Although Clostridium difficile is widely considered an antibiotic- and hospital-associated pathogen, recent evidence indicates that this is an insufficient depiction of the risks and reservoirs. A common thread that links all major risk factors of infection is their association with gastrointestinal disturbances, but this relationship to C. difficile colonization has never been tested directly. Here, we show that disturbances caused by diarrhoeal events trigger susceptibility to C. difficile colonization. Using survey data of the human gut microbiome, we detected C. difficile colonization and blooms in people recovering from food poisoning and Vibrio cholerae infections. Carriers remained colonized for year-long time scales and experienced highly variable patterns of C. difficile abundance, where increased shedding over short periods of 1–2 d interrupted week-long periods in which C. difficile was undetectable. Given that short shedding events were often linked to gastrointestinal disturbances, our results help explain why C. difficile is frequently detected as a co-infecting pathogen in patients with diarrhoea. To directly test the impact of diarrhoea on susceptibility to colonization, we developed a mouse model of variable disturbance intensity, which allowed us to monitor colonization in the absence of disease. As mice exposed to avirulent C. difficile spores ingested increasing quantities of laxatives, more individuals experienced C. difficile blooms. Our results indicate that the likelihood of colonization is highest in the days immediately following acute disturbances, suggesting that this could be an important window during which transmission could be interrupted and the incidence of infection lowered.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The publicly available data that we reanalysed here were generated by: David et al.30, European Nucleotide Archive under the accession number ERP006059; Hsaio et al.33, NCBI Short Read Archive (SRA) under the accession number PRJEB6358; Caporaso et al.36, MG-RAST under the accession number MG-RAST:4457768.3-4459735.3; Parker et al.34, SRA under accession number ERP022953; and The NIH Human Microbiome Project37 on NCBI under the accession number PRJNA43017. The amplicon sequencing data from the mouse model of varied disturbance intensity are available on the NCBI SRA under the accession number PRJNA507320. The genome of C. difficile BAA1801 is available on NCBI under the accession number PRJNA562328. All source data necessary to reproduce the figures presented in this manuscript are included in this article and its Supplementary information files. The microscope images used to calculate the cell and spore density are available online at Figshare (https://figshare.com/projects/Source_Data_for_Diarrheal_events_can_trigger_long-term_Clostridium_difficile_colonization_with_recurrent_blooms/72824).
Code availability
All custom computer code necessary to reproduce our results are available on GitHub (https://github.com/polzlab/VanInsberghe_2019_Cdiff_colonization).
References
Loo, V. G. et al. Host and pathogen factors for Clostridium difficile infection and colonization. N. Engl. J. Med. 365, 1693–1703 (2011).
Dubberke, E. R. & Olsen, M. A. Burden of Clostridium difficile on the healthcare system. Clin. Infect. Dis. 55, S88–S92 (2012).
Desai, K. et al. Epidemiological and economic burden of Clostridium difficile in the United States: estimates from a modeling approach. BMC Infect. Dis. 16, 303 (2016).
Kelly, C. R. et al. Effect of fecal microbiota transplantation on recurrence in multiply recurrent Clostridium difficile infection a randomized trial. Ann. Intern. Med. 165, 609–616 (2016).
Reveles, K. R., Lee, G. C., Boyd, N. K. & Frei, C. R. The rise in Clostridium difficile infection incidence among hospitalized adults in the United States: 2001-2010. Am. J. Infect. Control 42, 1028–1032 (2014).
Ray, A. J. et al. A multicenter randomized trial to determine the effect of an environmental disinfection intervention on the incidence of healthcare-associated Clostridium difficile infection. Infect. Control Hosp. Epidemiol. 38, 777–783 (2017).
Patton, A. et al. Impact of antimicrobial stewardship interventions on Clostridium difficile infection and clinical outcomes: segmented regression analyses. J. Antimicrob. Chemother. 73, 517–526 (2017).
Bruminhent, J. et al. Clostridium difficile colonization and disease in patients undergoing hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 20, 1329–1334 (2014).
Leffler, D. A. & Lamont, J. T. Clostridium difficile infection. N. Engl. J. Med. 372, 1539–1548 (2015).
Durovic, A., Widmer, A. F. & Tschudin-Sutter, S. New insights into transmission of Clostridium difficile infection—narrative review. Clin. Microbiol. Infect. 24, 483–492 (2018).
Crobach, M. J. T. et al. Understanding Clostridium difficile Colonization. Clin. Microbiol. Rev. 31, e00021-17 (2018).
Tschudin-Sutter, S. et al. Impact of toxigenic Clostridium difficile colonization on the risk of subsequent C. difficile infection in intensive care unit patients. Infect. Control Hosp. Epidemiol. 36, 1324–1329 (2015).
Eyre, D. W. et al. Diverse sources of C. difficile infection identified on whole-genome sequencing. N. Engl. J. Med. 369, 1195–1205 (2013).
Kociolek, L. K. et al. Clostridium difficile whole genome sequencing reveals limited transmission among symptomatic children: a single-center analysis. Clin. Infect. Dis. 67, 229–234 (2018).
Guh, A. Y. et al. Risk factors for community-associated Clostridium difficile infection in adults: a case-control study. Open Forum Infect. Dis. 4, ofx171 (2017).
McFarland, L. V., Surawicz, C. M. & Stamm, W. E. Risk factors for Clostridium difficile carriage and C. difficile-associated diarrhea in a cohort of hospitalized patients. J. Infect. Dis. 162, 678–684 (1990).
Dubberke, E. R. et al. Risk factors for acquisition and loss of Clostridium difficile colonization in hospitalized patients. Antimicrob. Agents Chemother. 59, 4533–4543 (2015).
Buffie, C. G. et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80, 62–73 (2012).
Brown, K. A., Khanafer, N., Daneman, N. & Fisman, D. N. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob. Agents Chemother. 57, 2326–2332 (2013).
Tedesco, F., Barton, R. & Alpers, D. Clindamycin-associated colitis: a prospective study. Ann. Intern. Med. 81, 429–433 (1974).
Weingarden, A. R. et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G310–G319 (2014).
Schubert, A. M., Sinani, H. & Schloss, P. D. Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. mBio 6, e00974-15 (2015).
Davis, M. Y., Zhang, H., Brannan, L. E., Carman, R. J. & Boone, J. H. Rapid change of fecal microbiome and disappearance of Clostridium difficile in a colonized infant after transition from breast milk to cow milk. Microbiome 4, 53 (2016).
Zackular, J. P. et al. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat. Med. 22, 1330–1334 (2016).
Seekatz, A. M., Rao, K., Santhosh, K. & Young, V. B. Dynamics of the fecal microbiome in patients with recurrent and nonrecurrent Clostridium difficile infection. Genome Med. 8, 47 (2016).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Daquigan, N., Seekatz, A. M., Greathouse, K. L., Young, V. B. & White, J. R. High-resolution profiling of the gut microbiome reveals the extent of Clostridium difficile burden. NPJ Biofilms Microbiomes 3, 35 (2017).
Vincent, C. et al. Reductions in intestinal Clostridiales precede the development of nosocomial Clostridium difficile infection. Microbiome 1, 18 (2013).
Wunderlin, T., Junier, T., Roussel-Delif, L., Jeanneret, N. & Junier, P. Endospore-enriched sequencing approach reveals unprecedented diversity of Firmicutes in sediments. Environ. Microbiol. Rep. 6, 631–639 (2014).
David, L. A. et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2015).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Vandeputte, D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017).
Hsiao, A. et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426 (2014).
Parker, E. P. K. et al. Changes in the intestinal microbiota following the administration of azithromycin in a randomised placebo-controlled trial among infants in south India. Sci. Rep. 7, 9168 (2017).
Jangi, S. & Lamont, J. T. Asymptomatic colonization by Clostridium difficile in infants: implications for disease in later life. J. Pediatr. Gastroenterol. Nutr. 51, 2–7 (2010).
Caporaso, J. G. et al. Moving pictures of the human microbiome. Genome Biol. 12, R50 (2011).
NIH HMP Working Group et al. The NIH Human Microbiome Project. Genome Res. 19, 2317–2323 (2009).
Aagaard, K. et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 27, 1012–1022 (2013).
Pashankar, D. S. & Bishop, W. P. Efficacy and optimal dose of daily polyethylene glycol 3350 for treatment of constipation and encopresis in children. J. Pediatr. 139, 428–432 (2001).
Kashyap, P. C. et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 144, 967–977 (2013).
Miao, J. et al. 16SPIP: a comprehensive analysis pipeline for rapid pathogen detection in clinical samples based on 16S metagenomic sequencing. BMC Bioinform. 18, 568 (2017).
van Prehn, J. et al. Diagnostic yield of repeat sampling with immunoassay, real-time PCR, and toxigenic culture for the detection of toxigenic Clostridium difficile in an epidemic and a non-epidemic setting. Eur. J. Clin. Microbiol. Infect. Dis. 34, 2325–2330 (2015).
Lawley, T. D. et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77, 3661–3669 (2009).
Murphy, C. N., Fowler, R. C., Iwen, P. C. & Fey, P. D. Evaluation of the BioFire FilmArray® GastrointestinalPanel in a Midwestern academic hospital. Eur. J. Clin. Microbiol. Infect. Dis. 36, 747–754 (2017).
Halligan, E. et al. Multiplex molecular testing for management of infectious gastroenteritis in a hospital setting: a comparative diagnostic and clinical utility study. Clin. Microbiol. Infect. 20, O460–O467 (2014).
Bartlett, J. G. Antibiotic-associated diarrhea. Clin. Infect. Dis. 15, 573–581 (1992).
Allegretti, J. R. et al. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment. Pharmacol. Ther. 43, 1142–1153 (2016).
Shim, J. K., Johnson, S., Samore, M. H., Bliss, D. Z. & Gerding, D. N. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet 351, 633–636 (1998).
Nagaro, K. J. et al. Nontoxigenic Clostridium difficile protects hamsters against challenge with historic and epidemic strains of toxigenic BI/NAP1/027 C. difficile. Antimicrob. Agents Chemother. 57, 5266–5270 (2013).
Eyre, D. W. et al. Asymptomatic Clostridium difficile colonisation and onward transmission. PLoS ONE 8, e78445 (2013).
Alasmari, F., Seiler, S. M., Hink, T., Burnham, C. A. D. & Dubberke, E. R. Prevalence and risk factors for asymptomatic Clostridium difficile carriage. Clin. Infect. Dis. 59, 216–222 (2014).
Scallan, E. et al. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17, 7–15 (2011).
Hartmann, M., Howes, C. G., Abarenkov, K., Mohn, W. W. & Nilsson, R. H. V-Xtractor: an open-source, high-throughput software tool to identify and extract hypervariable regions of small subunit (16S/18S) ribosomal RNA gene sequences. J. Microbiol. Methods 83, 250–253 (2010).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Pruesse, E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 (2007).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Lawley, T. D. et al. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J. Bacteriol. 191, 5377–5386 (2009).
Babiuk, L. A. & Paul, E. A. The use of fluorescein isothiocyanate in the determination of the bacterial biomass of grassland soil. Can. J. Microbiol. 16, 57–62 (1970).
Pau, G., Fuchs, F., Sklyar, O., Boutros, M. & Huber, W. EBImage—an R package for image processing with applications to cellular phenotypes. Bioinformatics 26, 979–981 (2010).
Preheim, S. P., Perrotta, A. R., Martin-Platero, A. M., Gupta, A. & Alm, E. J. Distribution-based clustering: using ecology to refine the operational taxonomic unit. Appl. Environ. Microbiol. 79, 6593–6603 (2013).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Untergasser, A. et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012).
Angiuoli, S. V. & Salzberg, S. L. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27, 334–342 (2011).
Acknowledgements
This work was supported by a grant from the Center for Microbiome Informatics and Therapeutics to M.F.P. D.V. was partially supported by a fellowship through the Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship-Doctoral. We thank I. Nachamkin (University of Pennsylvania) and N. Pollock (Harvard Medical School) for their comments, which helped improve this study.
Author information
Authors and Affiliations
Contributions
The concept was developed by D.V. and M.F.P. D.V. performed the 16S rRNA amplicon sequence data and C. difficile genomic analyses. D.V., M.F.P. and S.E. designed the mouse model of variable disturbance intensity and D.V. performed the experiment that utilizes this model with the help of B.V. J.A.E. designed the pipeline for analysing the 16S rRNA amplicon sequences from the mouse faecal samples. T.P. and S.E. performed the histological analyses. D.V. and M.F.P. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Representative histology of caecum, ileocecal junction, ileum and ascending colon from mice fed varying amounts of polyethylene glycol 3350 (PEG335) while being exposed to avirulent C. difficile spores.
All 40 mice from all treatment groups have comparable intestinal histology and show no evidence of typhlitis, enteritis or colitis. Tissues were collected ten days after treatment. Hematoxylin and Eosin. Bars= 100 m.
Extended Data Fig. 2 Increasing disturbance intensity increases the likelihood and magnitude C. difficile blooms in mice.
Four groups of ten C57BL/6J Mice were single housed in cages with grated bottoms. Non-toxinogenic C. difficile spores were suspended in drinking water at 104 spores/ml for one day prior, during, and two days following PEG3350 laxative treatment. Daily faecal samples were characterized using 16S rRNA PCR amplicon sequencing. The top panels summarize the family-level phylogenetic structure of the faecal microbial community over time, where different shades of the same colour represent different families in the same phyla; Firmicutes and Bacteroidetes families are depicted in green and blue, respectively. Lower panels show the relative abundance of C. difficile over time.
Extended Data Fig. 3 C. difficile blooms occur the day after treatment when cell density and stool consistency have returned to pre-treatment levels.
(a) The cell density of faecal samples was measured by direct microscopic counts of diluted samples stained with the fluorescent dye fluorescein isothiocyanate (FITC), which covalently binds the amine groups of proteins. Cell density was calculated using 25 images of random fields of the fixed and stained stool samples; center values represent the average cell density and error bars express the 95% confidence intervals of the densities observed for each sample. (b) To confirm the abundance of C. difficile measured using V4 16S rRNA amplicon sequencing (solid line), the abundance was also measured using a qPCR assay that specifically amplifies the rplP gene of C. difficile. (c) Representative stool samples from mice before, during, and after treatment are shown to illustrate how loose the stools of mice fed 150 mg/ml PEG3350 were at the end of treatment, but returned to normal by the following day. All ten mice in the highest PEG3350 treatment group experienced the same pattern of changes in stool consistency over time shown here.
Extended Data Fig. 4 Isolation of C. difficile colonies and genotype-specific colony PCR confirms that the inoculating strain is responsible for the blooms observed in mice treated with laxatives.
Faecal samples from the two mice that experienced the highest magnitude increase in C. difficile measured via 16S rRNA amplicon sequencing were anaerobically homogenized and two dilutions were plated on cycloserine-cefoxitin fructose agar (CCFA) in duplicate. (A) Only one colony morphology was detected from all samples; ground class texture and diffuse edges are consistent with C. difficile. (B) Colony forming unit (CFU) counts roughly reflect the abundance of C. difficile measured via 16S rRNA sequencing. (C) PCR confirms that all isolates isolated on CCFA plates are the same genotype as the inoculating strain (ATCC BAA1801). The C. difficile specific PCR assay amplifies the rplP locus present in all C. difficile strains, while the assay specific to the BAA1801 genotypic cluster amplifies a gene that is present in BAA1801 and its 19 closest sequenced relatives (differentiated by only 270 SNPs, genome-wide) but no other C. difficile lineage. C. difficile strain ATCC-9689 is a toxinogenic control that can be successfully amplified using the rplP PCR assay but not the assay specific to the BAA1801 genotypic cluster. All 72 isolated colonies from all plated dilutions that grew on the CCFA plates were genotyped with both the rplP and BAA1801 genotypic cluster specific PCR assays once.
Supplementary information
Supplementary Table 1
Summary of 16S rRNA amplicon sequencing data necessary to reproduce the analysis of the mouse model of variable disturbance intensity.
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 3
Statistical Source Data.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 4
Unprocessed DNA gel images.
Rights and permissions
About this article
Cite this article
VanInsberghe, D., Elsherbini, J.A., Varian, B. et al. Diarrhoeal events can trigger long-term Clostridium difficile colonization with recurrent blooms. Nat Microbiol 5, 642–650 (2020). https://doi.org/10.1038/s41564-020-0668-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-0668-2
This article is cited by
-
Is the microbiome the cause of irritable bowel syndrome and inflammatory bowel disease? Lessons to consider from odontology
International Journal of Colorectal Disease (2023)
-
Metabolic adaption to extracellular pyruvate triggers biofilm formation in Clostridioides difficile
The ISME Journal (2021)