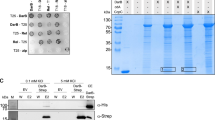
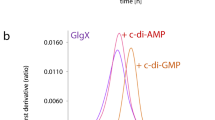
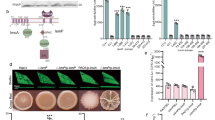
Abstract
Bacteria use small signalling molecules such as (p)ppGpp or c-di-GMP to tune their physiology in response to environmental changes. It remains unclear whether these regulatory networks operate independently or whether they interact to optimize bacterial growth and survival. We report that (p)ppGpp and c-di-GMP reciprocally regulate the growth of Caulobacter crescentus by converging on a single small-molecule-binding protein, SmbA. While c-di-GMP binding inhibits SmbA, (p)ppGpp competes for the same binding site to sustain SmbA activity. We demonstrate that (p)ppGpp specifically promotes Caulobacter growth on glucose, whereas c-di-GMP inhibits glucose consumption. We find that SmbA contributes to this metabolic switch and promotes growth on glucose by quenching the associated redox stress. The identification of an effector protein that acts as a central regulatory hub for two global second messengers opens up future studies on specific crosstalk between small-molecule-based regulatory networks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Data can be accessed via the Source Data files and Supplementary Tables. Mass spectrometry data are deposited on the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifiers PXD019846 and https://doi.org/10.6019/PXD019846. The Protein Data Bank accession numbers for the coordinates of the structures reported here are 6GS8 and 6GTM. Source data are provided with this paper.
References
Jenal, U., Reinders, A. & Lori, C. Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 (2017).
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T. & Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 (2015).
Davies, B. W., Bogard, R. W., Young, T. S. & Mekalanos, J. J. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370 (2012).
Dalebroux, Z. D. & Swanson, M. S. ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212 (2012).
Gentry, D. R., Hernandez, V. J., Nguyen, L. H., Jensen, D. B. & Cashel, M. Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. J. Bacteriol. 175, 7982–7989 (1993).
Milon, P. et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl Acad. Sci. USA 103, 13962–13967 (2006).
Wang, J. D., Sanders, G. M. & Grossman, A. D. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128, 865–875 (2007).
Maciag, M., Kochanowska, M., Lyzeń, R., Wegrzyn, G. & Szalewska-Pałasz, A. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid 63, 61–67 (2010).
Martins, D. et al. Superoxide dismutase activity confers (p)ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 115, 9797–9802 (2018).
Nguyen, D. et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986 (2011).
Schirmer, T. C-di-GMP synthesis: structural aspects of evolution, catalysis and regulation. J. Mol. Biol. 428, 3683–3701 (2016).
Römling, U., Galperin, M. Y. & Gomelsky, M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52 (2013).
Sudarsan, N. et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321, 411–413 (2008).
Tschowri, N. et al. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158, 1136–1147 (2014).
Lori, C. et al. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523, 236–239 (2015).
Kaczmarczyk, A. et al. Precise timing of transcription by c-di-GMP coordinates cell cycle and morphogenesis in Caulobacter. Nat. Commun. https://doi.org/10.1038/s41467-020-14585-6 (2020).
Ross, P. et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325, 279–281 (1987).
Hug, I., Deshpande, S., Sprecher, K. S., Pfohl, T. & Jenal, U. Second messenger-mediated tactile response by a bacterial rotary motor. Science 358, 531–534 (2017).
Boehm, A. et al. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141, 107–116 (2010).
Wang, B. et al. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150 (2019).
Mechold, U., Potrykus, K., Murphy, H., Murakami, K. S. & Cashel, M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 41, 6175–6189 (2013).
Rymer, R. U. et al. Binding mechanism of metal⋅NTP substrates and stringent-response alarmones to bacterial DnaG-type primases. Structure 20, 1478–1489 (2012).
Zhang, Y. E. et al. (p)ppGpp regulates a bacterial nucleosidase by an allosteric two-domain switch. Mol. Cell 74, 1239–1249.
Kanjee, U., Ogata, K. & Houry, W. A. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol. Microbiol. 85, 1029–1043 (2012).
Abel, S. et al. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the Caulobacter cell cycle. PLoS Genet. 9, e1003744 (2013).
Hallez, R., Delaby, M., Sanselicio, S. & Viollier, P. H. Hit the right spots: cell cycle control by phosphorylated guanosines in alphaproteobacteria. Nat. Rev. Microbiol. 15, 137–148 (2017).
Gonzalez, D. & Collier, J. Effects of (p)ppGpp on the progression of the cell cycle of Caulobacter crescentus. J. Bacteriol. 196, 2514–2525 (2014).
Nierman, W. C. et al. Complete genome sequence of Caulobacter crescentus. Proc. Natl Acad. Sci. USA 98, 4136–4141 (2001).
Marks, M. E. et al. The genetic basis of laboratory adaptation in Caulobacter crescentus. J. Bacteriol. 192, 3678–3688 (2010).
Kalyuzhnaya, M. G., Lidstrom, M. E. & Chistoserdova, L. Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J. 2, 696–706 (2008).
Nanadikar, M. S. et al. O2 affects mitochondrial functionality ex vivo. Redox Biol. 22, 101152 (2019).
Eruslanov, E. & Kusmartsev, S. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol. Biol. 594, 57–72 (2010).
Nesper, J., Reinders, A., Glatter, T., Schmidt, A. & Jenal, U. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J. Proteomics 75, 4874–4878 (2012).
Chou, S.-H. & Galperin, M. Y. Diversity of cyclic di-GMP-binding proteins and mechanisms. J. Bacteriol. 198, 32–46 (2016).
Mathis, R. & Ackermann, M. Response of single bacterial cells to stress gives rise to complex history dependence at the population level. Proc. Natl Acad. Sci. USA 113, 4224–4229 (2016).
Varik, V., Oliveira, S. R. A., Hauryliuk, V. & Tenson, T. HPLC-based quantification of bacterial housekeeping nucleotides and alarmone messengers ppGpp and pppGpp. Sci. Rep. 7, 11022 (2017).
Wang, Z. X. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 360, 111–114 (1995).
Hanson, G. T. et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279, 13044–13053 (2004).
Vilchèze, C., Hartman, T., Weinrick, B. & Jacobs, W. R. Mycobacterium tuberculosis is extraordinarily sensitive to killing by a vitamin C-induced Fenton reaction. Nat. Commun. 4, 1881 (2013).
Keyer, K. & Imlay, J. A. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl Acad. Sci. USA 93, 13635–13640 (1996).
Kalir, A. & Poljakoff-Mayber, A. Changes in activity of malate dehydrogenase, catalase, peroxidase and superoxide dismutase in leaves of Halimione portulacoides (L.) Aellen exposed to high sodium chloride concentrations. Ann. Bot. https://doi.org/10.1093/oxfordjournals.aob.a086002 (1981).
Smirnova, G. V., Muzyka, N. G. & Oktyabrsky, O. N. The role of antioxidant enzymes in response of Escherichia coli to osmotic upshift. FEMS Microbiol. Lett. 186, 209–213 (2000).
Narayanan, S., Janakiraman, B., Kumar, L. & Radhakrishnan, S. K. A cell cycle-controlled redox switch regulates the topoisomerase IV activity. Genes Dev. 29, 1175–1187 (2015).
Gardner, P. R. & Fridovich, I. Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J. Biol. Chem. 266, 1478–1483 (1991).
Seidler, N. W. GAPDH: Biological Properties and Diversity (Springer, 2012).
Whitfield, C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68 (2006).
Patel, K. B. et al. Functional characterization of UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferases of Escherichia coli and Caulobacter crescentus. J. Bacteriol. 194, 2646–2657 (2012).
Hershey, D. M. et al. Composition of the holdfast polysaccharide from Caulobacter crescentus. J. Bacteriol. 201, e00276 (2019).
Fiebig, A. et al. A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet. 10, e1004101 (2014).
Singh, R., Lemire, J., Mailloux, R. J. & Appanna, V. D. A novel strategy involved in anti-oxidative defense: the conversion of NADH into NADPH by a metabolic network. PLoS ONE 3, e2682 (2008).
Shimizu, K. & Matsuoka, Y. Redox rebalance against genetic perturbations and modulation of central carbon metabolism by the oxidative stress regulation. Biotechnol. Adv. 37, 107441 (2019).
Christen, M. et al. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328, 1295–1297 (2010).
Goemans, C. V. et al. An essential thioredoxin is involved in the control of the cell cycle in the bacterium Caulobacter crescentus. J. Biol. Chem. 293, 3839–3848 (2018).
Hartl, J. et al. Untargeted metabolomics links glutathione to bacterial cell cycle progression. Nat. Metab. 28, 153–166 (2020).
Zhang, Y., Zborníková, E., Rejman, D. & Gerdes, K. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. mBio 9, e02188-17 (2018).
Sévin, D. C., Stählin, J. N., Pollak, G. R., Kuehne, A. & Sauer, U. Global metabolic responses to salt stress in fifteen species. PLoS ONE 11, e0148888 (2016).
Leslie, A. G. W. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 (2006).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Potterton, L. et al. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr. D Struct. Biol. 74, 68–84 (2018).
Terwilliger, T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 (2009).
Terwilliger, T. C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Pervushin, K., Riek, R., Wider, G. & Wüthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl Acad. Sci. USA 94, 12366–12371 (1997).
Zähringer, F., Massa, C. & Schirmer, T. Efficient enzymatic production of the bacterial second messenger c-di-GMP by the diguanylate cyclase YdeH from E. coli. Appl. Biochem. Biotechnol. 163, 71–79 (2010).
Ahrné, E. et al. Evaluation and improvement of quantification accuracy in isobaric mass tag-based protein quantification experiments. J. Proteome Res. 15, 2537–2547 (2016).
Hartl, J., Kiefer, P., Meyer, F. & Vorholt, J. A. Longevity of major coenzymes allows minimal de novo synthesis in microorganisms. Nat. Microbiol. 2, 17073 (2017).
Wu, L. et al. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal. Biochem. 336, 164–171 (2005).
Kiefer, P., Delmotte, N. & Vorholt, J. A. Nanoscale ion-pair reversed-phase HPLC–MS for sensitive metabolome analysis. Anal. Chem. 83, 850–855 (2011).
Acknowledgements
We thank T. Sharpe (Biophysics Facility, Biozentrum, University of Basel), A. Schmidt and T. Bock (Proteomics Core Facility, Biozentrum, University of Basel), J. Bögli and S. Stefanova (FACS Core Facility, Biozentrum, University of Basel) for their technical guidance, F. Hamburger for cloning and strain construction, B. Lehtinen for help with the metabolite sampling, K. Sprecher and I. Hug for strain construction, and C. v. Arx for plasmids. We thank R. Hallez and S. Crosson for providing published plasmids. This work was supported by a European Research Council (ERC) Advanced Research Grant (grant no. 3222809 to U.J.), the Swiss National Science Foundation (grant nos. 310030B_147090 (to U.J.), 31003A_166652 (to T.S.) and 31003A_173094 (to J.A.V.)).
Author information
Authors and Affiliations
Contributions
Conceptualization: V.S., B.N.D., T.S. and U.J. Methodology: V.S., B.N.D, J.N., R.B., S.H., J.A.V. and J.H. Formal analysis: V.S., B.N.D., R.B., J.H., T.S. and U.J. Investigation: V.S., B.N.D., R.B., J.H., J.N., T.S. and U.J. Resources: U.J. and T.S. Writing of the original draft: V.S., B.N.D., T.S and U.J., with contributions from all other authors. Funding acquisition: T.S. and U.J.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Caulobacter growth on glucose is inversely regulated by (p)ppGpp and c-di-GMP.
(a) Growth of Caulobacter wild type and Δrel mutant inoculated from PYE stationary phase cultures into fresh M2G. For Δrel, adaptation periods and logarithmic growth are separated by a dashed red line. Growth curves were parameterized (adaptation phase t0, exponential growth rate k) by fitting a corresponding mathematical function to the data (see Methods). (b) and (c) Growth of Caulobacter wild type and mutant strains inoculated from logarithmically growing (b) and stationary phase cultures (c) into fresh M2G. Shown are mean of n = 3 biological replicates ± SD. (d) and (e) Ten-fold serial dilutions of Caulobacter wild type and mutant strains from exponentially growing (d) or stationary phase cultures (e) on solid MG2 agar plates. (f) Phase contrast microscopy images (left panel) and quantification of cell length (right panel) for Caulobacter wild type and Δrel mutant during adaptation and exponential growth phase. (g) Re-passaging experiments confirm the stability of the Δrel growth phenotype. The process of growth and re-plating is shown schematically. (h) and (i) Growth of Caulobacter rel + (h) and Δrel mutant (i) at gradually increasing c-di-GMP concentrations. (j) Growth of Caulobacter wild type and mutant strains inoculated from PYE stationary phase cultures into fresh M2X. (k) Uptake of 3H-glucose by Caulobacter wild type and mutant strains as indicated. The proton motive force inhibitor CCCP was used to block sugar uptake. (l) Transcription of the zwf operon as measure by lacZ reporter fusions in strains indicated grown in M2G. (m) Activity of the zwf promoter was determined as in (l) for the strains indicated. Pink and green dotted lines represent reporter activity in Caulobacter wild type grown in M2G and M2X, respectively. In panels (g-m), data represent the mean of n = 3 biological replicates ± SD.
Extended Data Fig. 2 Oxidative stress detection in Caulobacter.
(a) RSG fluorescence of Caulobacter wild type challenged with different levels of hydrogen peroxide for 10 min during exponential growth in M2G was measured by FACS. (b) Top: Schematic representation of Grx1-roGFP redox sensitive probe activation. Bottom: Exponentially growing Caulobacter wild type cultures expressing were treated with 10 mM hydrogen peroxide or 10 mM DTT and analysed by FACS. (c) The indicated Caulobacter strains expressing Grx1-roGFP were grown to the early adaptation phase in M2G and 405/488 nm excitation ratios were calculated as shown in (b). (d) Schematic of DCF-DA fluorescent probe activation. (e) ROS detection with the DCF-DA fluorescent probe in strains indicated during logarithmic growth and stationary phase in PYE or during adaptation in M2G. Shown are mean of n = 3 biological replicates ± SD.
Extended Data Fig. 3 Binding parameters of SmbA protein in complex with nucleotides.
(a-j) Isotherms representing binding of recombinant SmbA wild type or mutant proteins with c-di-GMP (a-d), ppGpp (e-h), pppGpp (i) and GMP (j) as measured by ITC. Please see Supplementary Table 2 for detailed experimental conditions and measured parameters.
Extended Data Fig. 4 Effects of second messenger binding to SmbA monitored by NMR spectroscopy.
(a) 2D [15N,1H]-TROSY spectrum of 160 μM [U-15N]-labelled SmbA in the apo state (magenta). (b) Spectral overlay of 160 μM apo SmbA (magenta) and 160 μM SmbA + 800 μM ppGpp (orange). (c) Spectral overlay of 160 μM apo SmbA (purple) and 160 μM SmbA + 300 μM c-di-GMP (light blue). (d) Spectral overlay of 160 μM SmbA + 300 μM c-di-GMP (blue) and 160 μM SmbA + 800 μM ppGpp (orange). All experiments were recorded in 30 mM HEPES pH 7.0, 250 mM NaCl, 5 mM MgCl2, 1 mM DTT measured at 20 °C on a 700 MHz spectrometer. The spectral region surrounded by the black box is enlarged in Fig. 2d.
Extended Data Fig. 5 Structural details of second messenger binding to SmbA.
(a) Cartoon representation of the (c-di-GMP)2 complex with full structural details of the ligands and interacting residues. H-bonds (length < 3.5 Å) and cation - π interactions are indicated by red and magenta lines, respectively. Note that R133*, which forms lateral H-bonds with O6 and N7 of g2 and cation - π interaction with g1, is donated by an adjacent protomer in the crystal packing (see Supplementary Fig. 7a). (b) Structure of the ppGpp complex. Note that the C-terminal carboxylate of a symmetry related protomer is forming a salt-bridge with R251 (see Supplementary Fig. 7b). (c) 2Fo-Fc omit maps contoured at 1.2 σ of (c-di-GMP)2 (left) and ppGpp (right) as bound to SmbA. (d) Fo-Fc omit maps contoured at 3.0 σ of (c-di-GMP)2 (left) and ppGpp (right).
Extended Data Fig. 6 (p)ppGpp and c-di-GMP inversely control SmbA activity to promote growth on glucose.
(a) Growth of Caulobacter wild type and different smbA mutants in M2G media. (b) Growth of Caulobacter wild type and mutants indicated in the rel + background diluted from stationary phase cultures (PYE) into fresh M2G containing 50 mM sodium chloride. The rcdG0::dgcZ strain harbours an IPTG-inducible (100 µM) copy of dgcZ on the chromosome. (c) Growth of Caulobacter wild type and Δrel mutant in M2X media supplemented with 50 mM sodium chloride. (d) Growth of Caulobacter Δrel mutants containing different smbA mutant alleles (as indicated) in M2G media supplemented with 30 mM sodium chloride. In panels (a-d), data represent the mean of n = 3 biological replicates ± SD. (e) SmbA specifically promotes growth on M2G. Caulobacter mutants smbA Q114A and smbA R143A harbouring plasmid pSA280 (Plac-dgcZ; 150 μM IPTG) were grown on minimal glucose (M2G), minimal xylose (M2X) and complex media (PYE) as indicated. Ratios of doubling times are indicated as a measure for the growth promoting role of SmbA under different conditions. Shown are the mean of n = 3 biological replicates ± SD. (f) Viability of Caulobacter Δrel mutants harbouring different smbA alleles was assayed by diluting cultures from stationary phase (PYE) onto solid MG2 agar plates. (g) Immunoblots of Caulobacter cell extracts (wild type and Δrel mutant) were stained using polyclonal anti-SmbA antisera. Cultures were grown as indicated in Fig. 1b and reached stationary at 15 hours. Similar results were obtained in three independent experiments.
Extended Data Fig. 7 (p)ppGpp and c-di-GMP control growth on glucose by balancing cellular redox state.
(a) Relative levels of radicals as measured by DCF fluorescence intensity in the strains indicated. Fluorescence was scored in exponentially growing cells in M2X (left bar) or in M2X supplemented with 10 mM ascorbic acid (right bar). Dashed lines indicate the levels of DCF fluorescence measured in Caulobacter wild type treated with 10 mM ascorbic acid (green) or untreated (black). (b–e) Growth of Caulobacter wild type and indicated mutant strains. Cells were diluted from cultures growing exponentially in PYE to fresh M2G or M2X with or without 10 mM ascorbic acid as indicated. In panels (a-e), data represent the mean of n = 3 biological replicates ± SD. (f) Flow cytometry analysis of Caulobacter wild type and mutant cells grown on M2X. Cells were sampled after growth in M2X for 15 h as indicated in (d) and (e) and samples were incubated with RSG for 20 min before FACS analysis. (g) Caulobacter cell cycle progression is affected by (p)ppGpp and SmbA. The optical density of synchronized populations of wild type and mutant strains was scored during one cell cycle as indicated. (h) Immunoblot analysis of synchronized populations indicated in (g) using polyclonal anti-CtrA antisera. CtrA protein levels are used as a marker for cell cycle progression (indicated for Caulobacter wild type above panels in g and h). Similar results were obtained in three independent experiments.
Extended Data Fig. 8 SmbA regulates Caulobacter surface attachment.
(a) Fraction of Caulobacter wild type and smbA mutants with visible holdfast. (b) Representative microscopy images (overlay of phase contrast and fluorescent GFP channels) of cultures with stained holdfast (indicated by red arrows) used for calculations described above. For each strain, similar results were obtained from three independent cultures. (c) Transcription of hfiA in Caulobacter wild type and mutant strains as indicated. A lacZ transcriptional reporter was used to determine hfiA promoter strength. β-galactosidase activities were determined in strains containing different smbA alleles on the chromosome. A staR deletion mutant was used as a control. (d) Attachment of hfiA deletion strains harbouring different smbA alleles as measured by crystal violet stain. In panels (a, c-d), data represent the mean of n = 3 biological replicates ± SD.
Supplementary information
Supplementary information
Supplementary Figs. 1–5, Supplementary Tables 2, 4–7 and Supplementary Methods.
Supplementary Table 1
List of all peptides identified from the CCMS for cdG effector screen (S1) and mass spectrometry-based proteome analysis (S3).
Supplementary Data 1
Statistical source data for Supplementary Fig. 4.
Source data
Source Data Fig. 1
Source data.
Source Data Fig. 2
Original autoradiographs from three independent experiments.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 5
Source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 6
Representative original unmodified gels.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Representative original unmodified gels.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Shyp, V., Dubey, B.N., Böhm, R. et al. Reciprocal growth control by competitive binding of nucleotide second messengers to a metabolic switch in Caulobacter crescentus. Nat Microbiol 6, 59–72 (2021). https://doi.org/10.1038/s41564-020-00809-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-00809-4
This article is cited by
-
c-di-GMP inhibits the DNA binding activity of H-NS in Salmonella
Nature Communications (2023)
-
Mutant structure of metabolic switch protein in complex with monomeric c-di-GMP reveals a potential mechanism of protein-mediated ligand dimerization
Scientific Reports (2023)
-
cAMP and c-di-GMP synergistically support biofilm maintenance through the direct interaction of their effectors
Nature Communications (2022)
-
Autoinducer-2 and bile salts induce c-di-GMP synthesis to repress the T3SS via a T3SS chaperone
Nature Communications (2022)
-
Crosstalking second messengers
Nature Microbiology (2020)