Abstract
Selective recruitment and concentration of signalling proteins within membraneless compartments is a ubiquitous mechanism for subcellular organization1,2,3. The dynamic flow of molecules into and out of these compartments occurs on faster timescales than for membrane-enclosed organelles, presenting a possible mechanism to control spatial patterning within cells. Here, we combine single-molecule tracking and super-resolution microscopy, light-induced subcellular localization, reaction-diffusion modelling and a spatially resolved promoter activation assay to study signal exchange in and out of the 200 nm cytoplasmic pole-organizing protein popZ (PopZ) microdomain at the cell pole of the asymmetrically dividing bacterium Caulobacter crescentus4,5,6,7,8. Two phospho-signalling proteins, the transmembrane histidine kinase CckA and the cytoplasmic phosphotransferase ChpT, provide the only phosphate source for the cell fate-determining transcription factor CtrA9,10,11,12,13,14,15,16,17,18. We find that all three proteins exhibit restricted rates of entry into and escape from the microdomain as well as enhanced phospho-signalling within, leading to a submicron gradient of activated CtrA-P19 that is stable and sublinear. Entry into the microdomain is selective for cytosolic proteins and requires a binding pathway to PopZ. Our work demonstrates how nanoscale protein assemblies can modulate signal propagation with fine spatial resolution, and that in Caulobacter, this modulation serves to reinforce asymmetry and differential cell fate of the two daughter cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The BacTRIP sequencing data for the mapping, transcription and normalization datasets have been uploaded to the National Center for Biotechnology Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/ under accession no. PRJNA595753. The lists of plasmids and strains generated as part of this study are available in Supplementary Tables 2 and 3. A complete list of parameters used for modelling the CtrA activation pathway is available in Supplementary Table 7. Source data for most panels in the main and Extended Data figures is available online in the Source Data. The remaining data supporting the findings of this study are available from the corresponding author on request.
Code availability
The code that supports the findings of this study, including the analysis of single-molecule tracks, reaction-diffusion model of CtrA activation pathway and analysis of the BacTRIP data are available from the corresponding author on request.
References
Shapiro, L., McAdams, H. H. & Losick, R. Why and how bacteria localize proteins. Science 326, 1225–1228 (2009).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018).
Bowman, G. R. et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol. Microbiol. 76, 173–189 (2010).
Bowman, G. R. et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134, 945–955 (2008).
Ebersbach, G., Briegel, A., Jensen, G. J. & Jacobs-Wagner, C. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134, 956–968 (2008).
Holmes, J. A. et al. Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles. Proc. Natl Acad. Sci. USA 113, 12490–12495 (2016).
Bergé, M. & Viollier, P. H. End-in-sight: cell polarization by the polygamic organizer PopZ. Trends Microbiol. 26, 363–375 (2018).
Jacobs, C., Ausmees, N., Cordwell, S. J., Shapiro, L. & Laub, M. T. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol. Microbiol. 47, 1279–1290 (2003).
Biondi, E. G. et al. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444, 899–904 (2006).
Chen, Y. E., Tsokos, C. G., Biondi, E. G., Perchuk, B. S. & Laub, M. T. Dynamics of two phosphorelays controlling cell cycle progression in Caulobacter crescentus. J. Bacteriol. 191, 7417–7429 (2009).
Angelastro, P. S., Sliusarenko, O. & Jacobs-Wagner, C. Polar localization of the CckA histidine kinase and cell cycle periodicity of the essential master regulator CtrA in Caulobacter crescentus. J. Bacteriol. 192, 539–552 (2010).
Iniesta, A. A., Hillson, N. J. & Shapiro, L. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc. Natl Acad. Sci. USA 107, 7012–7017 (2010).
Tsokos, C. G. & Laub, M. T. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr. Opin. Microbiol. 15, 744–750 (2012).
Blair, J. A. et al. Branched signal wiring of an essential bacterial cell-cycle phosphotransfer protein. Structure 21, 1590–1601 (2013).
Lori, C. et al. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523, 236–239 (2015).
Lasker, K., Mann, T. H. & Shapiro, L. An intracellular compass spatially coordinates cell cycle modules in Caulobacter crescentus. Curr. Opin. Microbiol. 33, 131–139 (2016).
Mann, T. H., Seth Childers, W., Blair, J. A., Eckart, M. R. & Shapiro, L. A cell cycle kinase with tandem sensory PAS domains integrates cell fate cues. Nat. Commun. 7, 11454 (2016).
Chen, Y. E. et al. Spatial gradient of protein phosphorylation underlies replicative asymmetry in a bacterium. Proc. Natl Acad. Sci. USA 108, 1052–1057 (2011).
Ptacin, J. L. et al. Bacterial scaffold directs pole-specific centromere segregation. Proc. Natl Acad. Sci. USA 111, E2046–E2055 (2014).
Gahlmann, A. et al. Quantitative multicolor subdiffraction imaging of bacterial protein ultrastructures in three dimensions. Nano Lett. 13, 987–993 (2013).
Tsokos, C. G., Perchuk, B. S. & Laub, M. T. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev. Cell 20, 329–341 (2011).
Mann, T. H. & Shapiro, L. Integration of cell cycle signals by multi-PAS domain kinases. Proc. Natl Acad. Sci. USA 115, E7166–E7173 (2018).
Childers, W. S. et al. Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS Biol. 12, e1001979 (2014).
Levskaya, A., Weiner, O. D., Lim, W. A. & Voigt, C. A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009).
Guntas, G. et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl Acad. Sci. USA 112, 112–117 (2015).
Dubey, B. N. et al. Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking. Sci. Adv. 2, e1600823 (2016).
Brown, G. C. & Kholodenko, B. N. Spatial gradients of cellular phospho-proteins. FEBS Lett. 457, 452–454 (1999).
Panis, G., Murray, S. R. & Viollier, P. H. Versatility of global transcriptional regulators in alpha-Proteobacteria: from essential cell cycle control to ancillary functions. FEMS Microbiol. Rev. 39, 120–133 (2015).
Zhou, B. et al. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet. 11, e1004831 (2015).
Collier, J., McAdams, H. H. & Shapiro, L. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc. Natl Acad. Sci. USA 104, 17111–17116 (2007).
Narayanan, S., Kumar, L. & Radhakrishnan, S. K. Sensory domain of the cell cycle kinase CckA regulates the differential DNA binding of the master regulator CtrA in Caulobacter crescentus. Biochim. Biophys. Acta Gene Regul. Mech. 1861, 952–961 (2018).
Viollier, P. H. et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl Acad. Sci. USA 101, 9257–9262 (2004).
Heid, C. A., Stevens, J., Livak, K. J. & Williams, P. M. Real time quantitative PCR. Genome Res. 6, 986–994 (1996).
Domian, I. J., Quon, K. C. & Shapiro, L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90, 415–424 (1997).
Quon, K. C., Marczynski, G. T. & Shapiro, L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84, 83–93 (1996).
Iniesta, A. A., McGrath, P. T., Reisenauer, A., McAdams, H. H. & Shapiro, L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl Acad. Sci. USA 103, 10935–10940 (2006).
Ryan, K. R., Huntwork, S. & Shapiro, L. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc. Natl Acad. Sci. USA 101, 7415–7420 (2004).
Akhtar, W. et al. Using TRIP for genome-wide position effect analysis in cultured cells. Nat. Protoc. 9, 1255–1281 (2014).
McAdams, H. H. & Shapiro, L. System-level design of bacterial cell cycle control. FEBS Lett. 583, 3984–3991 (2009).
Biondi, E. G. et al. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol. Microbiol. 59, 386–401 (2006).
Fumeaux, C. et al. Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nat. Commun. 5, 4081 (2014).
Tan, M. H., Kozdon, J. B., Shen, X., Shapiro, L. & McAdams, H. H. An essential transcription factor, SciP, enhances robustness of Caulobacter cell cycle regulation. Proc. Natl Acad. Sci. USA 107, 18985–18990 (2010).
Gora, K. G. et al. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol. Cell 39, 455–467 (2010).
Haakonsen, D. L., Yuan, A. H. & Laub, M. T. The bacterial cell cycle regulator GcrA is a σ70 cofactor that drives gene expression from a subset of methylated promoters. Genes Dev. 29, 2272–2286 (2015).
Janakiraman, B., Mignolet, J., Narayanan, S., Viollier, P. H. & Radhakrishnan, S. K. In-phase oscillation of global regulons is orchestrated by a pole-specific organizer. Proc. Natl Acad. Sci. USA 113, 12550–12555 (2016).
Reisenauer, A. & Shapiro, L. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 21, 4969–4977 (2002).
Gonzalez, D., Kozdon, J. B., McAdams, H. H., Shapiro, L. & Collier, J. The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res. 42, 3720–3735 (2014).
Laub, M. T., McAdams, H. H., Feldblyum, T., Fraser, C. M. & Shapiro, L. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290, 2144–2148 (2000).
Laub, M. T., Chen, S. L., Shapiro, L. & McAdams, H. H. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl Acad. Sci. USA 99, 4632–4637 (2002).
Quon, K. C., Yang, B., Domian, I. J., Shapiro, L. & Marczynski, G. T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl Acad. Sci. USA 95, 120–125 (1998).
Jenal, U. The role of proteolysis in the Caulobacter crescentus cell cycle and development. Res. Microbiol. 160, 687–695 (2009).
Rudner, D. Z. & Losick, R. Protein subcellular localization in bacteria. Cold Spring Harb. Perspect. Biol. 2, a000307 (2010).
Surovtsev, I. V. & Jacobs-Wagner, C. Subcellular organization: a critical feature of bacterial cell replication. Cell 172, 1271–1293 (2018).
Folkmann, A. W. & Seydoux, G. Single-molecule study reveals the frenetic lives of proteins in gradients. Proc. Natl Acad. Sci. USA 115, 9336–9338 (2018).
Li, L. et al. Real-time imaging of Huntingtin aggregates diverting target search and gene transcription. eLife 5, e17056 (2016).
Bianchi, F. et al. Steric exclusion and protein conformation determine the localization of plasma membrane transporters. Nat. Commun. 9, 501 (2018).
Lu, H. et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 (2018).
Wu, X. et al. RIM and RIM-BP form presynaptic active-zone-like condensates via phase separation. Mol. Cell 73, 971–984 (2019).
Case, L. B., Ditlev, J. A. & Rosen, M. K. Regulation of transmembrane signaling by phase separation. Annu. Rev. Biophys. 48, 465–494 (2019).
Huang, W. Y. C. et al. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103 (2019).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Thanbichler, M., Iniesta, A. A. & Shapiro, L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35, e137 (2007).
Toettcher, J. E., Weiner, O. D. & Lim, W. A. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434 (2013).
Poindexter, J. S. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28, 231–295 (1964).
Ely, B. Genetics of Caulobacter crescentus. Methods Enzymol. 204, 372–384 (1991).
Tsai, J. W. & Alley, M. R. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J. Bacteriol. 183, 5001–5007 (2001).
Schrader, J. M. & Shapiro, L. Synchronization of Caulobacter crescentus for investigation of the bacterial cell cycle. J. Vis. Exp. https://doi.org/10.3791/52633 (2015).
Perez, A. M. et al. A localized complex of two protein oligomers controls the orientation of cell polarity. mBio 8, e02238–16 (2017).
Lew, M. D., Backlund, M. P. & Moerner, W. E. Rotational mobility of single molecules affects localization accuracy in super-resolution fluorescence microscopy. Nano Lett. 13, 3967–3972 (2013).
von Diezmann, A., Lee, M. Y., Lew, M. D. & Moerner, W. E. Correcting field-dependent aberrations with nanoscale accuracy in three-dimensional single-molecule localization microscopy. Optica 2, 985–993 (2015).
Ovesný, M., Křížek, P., Borkovec, J., Svindrych, Z. & Hagen, G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45 (2001).
Reisenauer, A., Kahng, L. S., McCollum, S. & Shapiro, L. Bacterial DNA methylation: a cell cycle regulator? J. Bacteriol. 181, 5135–5139 (1999).
Siam, R. & Marczynski, G. T. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 19, 1138–1147 (2000).
Iyer-Biswas, S. et al. Scaling laws governing stochastic growth and division of single bacterial cells. Proc. Natl Acad. Sci. USA 111, 15912–15917 (2014).
Wright, C. S. et al. Intergenerational continuity of cell shape dynamics in Caulobacter crescentus. Sci. Rep. 5, 9155 (2015).
Brassinga, A. K. & Marczynski, G. T. Replication intermediate analysis confirms that chromosomal replication origin initiates from an unusual intergenic region in Caulobacter crescentus. Nucleic Acids Res. 29, 4441–4451 (2001).
Dingwall, A. & Shapiro, L. Rate, origin, and bidirectionality of Caulobacter chromosome replication as determined by pulsed-field gel electrophoresis. Proc. Natl Acad. Sci. USA 86, 119–123 (1989).
Umbarger, M. A. et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol. Cell 44, 252–264 (2011).
Le, T. B., Imakaev, M. V., Mirny, L. A. & Laub, M. T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734 (2013).
Kozdon, J. B. et al. Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. Proc. Natl Acad. Sci. USA 110, E4658–E4667 (2013).
Schrader, J. M. et al. The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet. 10, e1004463 (2014).
Spencer, W., Siam, R., Ouimet, M. C., Bastedo, D. P. & Marczynski, G. T. CtrA, a global response regulator, uses a distinct second category of weak DNA binding sites for cell cycle transcription control in Caulobacter crescentus. J. Bacteriol. 191, 5458–5470 (2009).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Acknowledgements
We thank J.W. Kern for help with plasmid design and construction, M.D. Melfi for providing the RT–qPCR protocol, A. Lovell and M.R. Eckart at the Stanford Protein and Nucleic Acid Facility for support in using MySeq and RT–qPCR and for running the surface plasmon resonance experiments, J.M. Schrader for sharing unpublished data on mRNA half-life in Caulobacter, A. Olson and the Stanford Neuroscience Microscopy Service (supported by grant no. NIH NS069375) for providing equipment for and assistance with the photobleaching experiments and A.H. Squires for critical feedback on the manuscript. We also thank H.H. McAdams for helpful discussions on the modelling of signal transduction and all members of the Shapiro and Moerner laboratories for helpful discussions throughout the project. We acknowledge support from the Gordon and Betty Moore Function (award no. GBMF 2550.03) to the Life Sciences Research Foundation (to K.L.), the Weizmann Institute of Science National Postdoctoral Award Program for Advancing Women in Science (to K.L.) and from the National Institute of General Medical Sciences of the National Institutes of Health under award nos. T32GM007276 to T.H.M, R01-GM086196 to W.E.M. and L.S., R35-GM118067 to W.E.M. and R35-GM118071 to L.S. L.S. is a Chan Zuckerberg Biohub Investigator. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
This study was conceived by K.L. and L.v.D. The study was designed by K.L., L.v.D., W.E.M. and L.S. The single-molecule experiments and Monte Carlo diffusion simulations were performed and analysed by L.v.D. The diffraction-limited imaging and photobleaching experiments were performed and analysed by K.L. and L.v.D. The qPCR, surface plasmon resonance and western blot data were collected by K.L. and D.G.A. and analysed by K.L. The integrative reaction-diffusion modelling was designed and interpreted by K.L. and L.v.D., and implemented and performed by K.L. BacTRIP was designed, implemented and analysed by K.L. and X.Z. The paper was written by K.L., L.v.D., T.H.M., W.E.M. and L.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
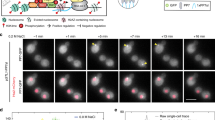
Extended Data Fig. 1 Diffraction-limited profiles of CckA-eYFP, ChpT-eYFP, CtrA-eYFP-14 localization, ChpT/PopZ interaction, and PopZ-eYFP dynamics.
a. Fluorescent constructs used to image the co-localization of CckA, ChpT, and CtrA with respect to PopZ (cf. Supplementary Table 1). pamcherry-popZ, cckA-eyfp and chpT-eyfp are integrated at the native promotor/locus. CtrA-eYFP-14 (sandwich fusion with C-terminal degradation tag) is driven by a PxylX promoter and is expressed from a high copy plasmid. b. CtrA-eYFP-14 has similar cell cycle behavior as CtrA. Shown is a time-resolved western blot of CtrA protein expressed from CtrA promoter and CtrA-eYFP-14 protein expressed from a vanillate-regulated promoter probed with anti-CtrA antibody. The KL6039 strain was induced with 1 mM vanillate for 1.5 hours prior to induction and during recovery. Data are representative of three biological replicates. c. Diffraction-limited images of predivisional Caulobacter cells expressing CckA-eYFP, ChpT-eYFP, or CtrA-eYFP-14 (top row) and PAmCherry-PopZ (middle row); new poles marked with a white arrow. Scale bars: 2 µm. Fluorescence intensity profiles, including percentage of CckA, ChpT, or CtrA signal at new and old poles, shown along normalized cell length (n = 55, 129, 143 cells respectively). d. Purified WT ChpT binds directly to PopZ, as measured by surface plasmon resonance (left), while purified CtrA does not (right). n equals three biologically independent samples. e. Recovery following targeted photobleaching of a portion of an extended PopZ microdomain. ΔpopZ cells expressing popZ-eyfp from a high copy plasmid were imaged for many frames of laser scanning confocal microscopy following targeted photobleaching with high-intensity 514 nm laser light. (left) The fraction of signal in the bleached region compared to signal in the entire microdomain is shown in red, with exponential fit in black. Shown is the mean ± s.d.; n equal to 6 cells. (right) Representative field of view from the PopZ FRAP experiment. A bleached cell and its recovery dynamics is shown next to an unbleached cell. Scale bars: 2 μm.
Extended Data Fig. 2 CckA dynamics within the poles.
a-c, simulations of membrane motion (cf. Supplementary Note 1.9). a. A single Brownian trajectory on the cell surface. The 2D projection of the trajectory exhibits erroneously low diffusivity at the poles and equator of the cell, where 3D information is lost. b. Left: mean-squared-displacement analysis of 3D diffusion accurately infers the diffusion coefficient of motion on the cell membrane, while restriction to 2D measurements induces systematic errors. Right: comparison of D values estimated from subtrajectories inside and outside the poles. Relative D underestimation inside the poles was ≤ 8% for the measured values of polar D for CckA-eYFP (0.02-0.04 µm2/s). c. Experimental approach of 3D tracking using the DH-PSF. Left, 4 second CckA-eYFP trajectory overlaid on brightfield image of a Caulobacter cell. Middle, translation and rotation in the double-helix PSF is used to detect 2D and 3D motion. Right, the corresponding 3D trajectory along the circumference of the cell. Scale bar: 500 nm. d. Caulobacter cells expressing CckA-eYFP imaged with confocal microscopy following targeted photobleaching. Signal at the old and new pole regions is defined as the fraction of total cell fluorescence (upper plot, errorbars show ± 95% CI). After 10 minutes, the initial ratio of old to new pole signal is restored (lower plot). Average of n = 6 cells. Scale bar: 2 μm. e. Fluorescence decay in cells with photobleaching only from imaging, (n = 9, cyan), cells that were uniformly bleached with the photobleaching laser (n = 3, red), and with targeted photobleaching at the old poles (n = 6, magenta) (same statistics as d.). f-g. data shown in d. if using a correction factor from ‘unbleached’ or ‘completely bleached’ cells. Regardless of the correction factor used, new poles exhibit loss and old poles exhibit recovery. Errorbars: 95% CI. h. Representative images of cells showing the StpX-mChy marker used to identify stalks, that is the old pole, and the conditions necessary for inclusion as ‘predivisional,’ as consistently observed in all cells. Scale bar: 2 μm. i. Predicted concentrations from reaction-diffusion simulations of CckA motion with fast and slow unbinding to the microdomain overlaid with data of (d). CckA molecules at the old pole were ‘bleached’ by marking the molecules as dark CckA molecules.
Extended Data Fig. 3 Super-resolved distribution of PopZ, CckA, ChpT, and CtrA within the poles.
a. Single-molecule distributions of CckA and PopZ localizations averaged from old poles (n = 29 poles) and new poles (n = 13) of predivisional cells. Blue arrows show the measured diameters of CckA. Scale bar: 200 nm. b. Projection of all single-molecule localizations of CckA and PopZ in the new pole, shown facing down the cell axis (531 and 477 localizations, respectively). Top and right: 1D histogram of CckA and PopZ with 25 nm bins. Centre: mean. The half widths at half maximum (HWHM) are calculated for CckA from the standard deviations of molecule positions along the Y and Z axes, with magnitudes shown by blue arrows (error: 95% CI from localization resampling). c. 1D profiles and 2D histograms (25 nm bin size) of CtrA, ChpT, and PopZ localizations averaged with respect to PopZ (60, 27, and 27 cells respectively for each reconstruction). Scale bar: 200 nm.
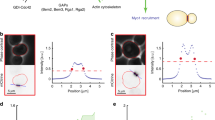
Extended Data Fig. 4 Selective polar permeability and motion of cytoplasmic proteins.
a-c. Simulations of freely diffusing cytoplasmic molecules with D = 1.8 μm2/s are observed in the poles (Supplementary Note 2.9). A step size of 0.5 ms was used, and to match experimental conditions, trajectories were reconstructed at 20 ms sampling rate (mimicking motion blur) and 40 nm localization error. a. The XY components of a single 3D trajectory, corresponding to localizations of a single freely-diffusing molecule. b. The histogram of localizations from 1,000 simulated 2-second trajectories, with poles defined as extending200 and 250 nm away from the tip (consistent with definitions used for experimental data) marked. c. 1D profile of histogram shown in (b). Approximately 4.1% and 5.5% of localizations would be expected to appear within the new and old poles (orange), respectively, if molecular motion were unobstructed. d. White light image and 2D histogram (40 nm bin size) of fPIF-eYFP and PAmCherry-PopZ single-molecule localizations. Scale bar: 2 μm. Consistent in 16 cells analyzed (Fig. 2b). e. White light image and 2D histogram (40 nm bin size) of eYFP and PAmCherry-PopZ single-molecule localizations. Scale bar: 2 μm. Consistent in 17 cells analyzed (Fig. 2b). f. 1D MSD values in the body of predivisional Caulobacter cells as shown in Fig. 1h, including eYFP and fPIF. 95% CI shown. g. Fitting the MSD data to an equation for anomalous diffusion was not able to precisely specify D and α due to the short track durations available. Points: fits values of D and α generated from 200 resamples of CtrA and ChpT tracks either from the cell body or from the poles. Blue lines mark combinations of D and α that would yield apparent diffusion coefficients Dapp for a given short-time approximation, calculated as \(D_{app} = D\tau ^{1 - \alpha }\) for Dapp = 1.8 μm2/s and τ = 20 ms (the frame integration time, right line) or τ = 100 ms (the maximum time included in the fit, left line). The trajectories measured are consistent with a short-time value of Dapp = 1.8 μm2/s and suggest that α < 0.7. In contrast, diffusion within the poles appeared relatively Brownian, with 0.75 ≤ α ≤ 0.9. The MSD curve of ChpT at the poles cannot be accurately fit due to confinement effects.
Extended Data Fig. 5 Correlative imaging of a membrane-bound, pole-insensitive construct and PopZ.
a. Diffraction-limited epifluorescence data of the FixL(TM-PAS)-eYFP construct (the transmembrane and PAS domains of Caulobacter FixL fused to eYFP) and PAmCherryPopZ. Overlap of PopZ and FixL(TM-PAS)-eYFP at the poles was consistently observed in 20 cells. Scale bar: 2 μm b. Brightfield image and 2-color histogram of single-molecule localizations of FixL(TM-PAS)-eYFP and PAmCherry-PopZ. Scale bar: 2 μm. c. A selection of five single-molecule tracks of the upper cell in b., shown relative to the PAmCherry-PopZ super-resolution reconstruction (red density). Detailed single-molecule analysis showed consistent entry into the membrane proximal to PopZ for the 4 cells analyzed. Scale bar: 200 nm.
Extended Data Fig. 6 Setup of the reaction-diffusion simulation of CtrA activation pathway.
a. A simplified schematic of the reaction and diffusion events used in our CtrA activation model. Dotted red arrows indicate CckA, ChpT, and CckA binding to either the new pole (NP) or the old pole (OP) species. The width of the arrow represents the strength of binding, with thicker arrows for tighter binding. The colored edges out of CckA (blue), ChpT (orange), and CtrA (green) boxes indicate diffusion inside the pole and in the body of the cell. The length of the arrow represents the rate of diffusion, with shorter arrows representing slower diffusion. Phosphotransfer reactions are shown in black. Each arrow is labeled with a reaction number from 1 to 39. Information on each reaction is found in Supplementary Table 7. b. A complete schematic of the phosphotransfer reactions including binding and transfer events. Reaction numbers match the simplified schematic in (a). c. Concentrations of each molecular species in the system were modeled as a function of position along the long axis of the cell and time. The governing partial differential equation (PDE) for each species was approximated as a set of ordinary differential equations (ODEs). The spatial dimension of a 4 μm cell was discretized into n = 80 bins, each representing a 0.05 μm Δx section of the cell. Each species is broken into n sub-species, one per bin. The simulation advances in 0.1 second steps. We use the second-order center difference formula to approximate the Laplacian operator and convert each PDE into a set of ODEs (Supplementary Note 2.2, equation S7). d. Simulation results with wildtype parameters (Fig. 3a) showing steady-state protein concentrations for CckA (blue), ChpT (orange), and CtrA (green) as a function of distance from the new pole in the predivisional cell.
Extended Data Fig. 7 Reaction-diffusion simulation of the CtrA activation pathway.
a. Global analysis of the effect of diffusion coefficients and reaction rates on CtrA~P amplitude and distribution. Sensitivity of CtrA~P amplitude (upper heat map) and CtrA~P distribution (lower heat map) to changes in (i) the rate of CckA, ChpT, and CtrA binding to either the new pole or old pole microdomains, (ii) diffusion coefficients of either CckA, ChpT, or CtrA, and (iii) phosphotransfer rates. Each heat map rectangle specifies one parameter (for example top/left rectangle specifies sensitivity to changes in the binding rate between CckA and the new pole microdomain). The color of the rectangle indicates the normalized sensitivity value (from low sensitivity (blue) to high sensitivity (red)) to changes in parameter value. b. Detailed results for changes in 8 parameters. For each parameter the fraction of total CtrA~P (top) and distribution of CtrA~P along the long axis of the cell (bottom) is shown for steady-state. Gray arrows signify the increasing value of the parameter. c. Contribution of the rate of phosphotrasfer from CckA to ChpT on the distribution of CtrA~P. We simulated the distribution of CtrA~P as a function of three different phosphorylation rates of CckA by ChpT: 10/s forward and 1/s back (blue), 100/s forward and 10/s back (green), and 100/s forward and 1/s back (brown). We varied the binding affinity between ChpT and PopZ between 0 and 10 μM, as well as the auto-phosphorylation rate of CckA: slow by setting CckA kHalf to 150 (left) as well as fast by setting the kHalf to 5 (right).
Extended Data Fig. 8 Diffraction-limited profiles of CckA-eYFP, ChpT-eYFP, CtrA-eYFP-14 localization and expression of CtrA~P-regulated genes in PopZ overexpression background.
a. Overexpression of PopZ does not affect CckA and ChpT abundance but does result in a reduction in CtrA abundance. Mixed population western blot of CckA-eYFP protein expressed from the cckA promoter, ChpT-eYFP expressed from the chpT promoter, and CtrA-14-eYFP protein expressed from a vanillate-regulated promoter. CckA-eYFP and ChpT-eYFP were probed with anti-YFP antibody while CtrA-14-eYFP was probed with anti-CtrA antibody. The protein abundance was measured in both wildtype (I) and PopZ overexpression (II) conditions. n equals three biologically independent experiments repeated with similar results. b. Diffraction-limited images of predivisional Caulobacter cells expressing CckA-eYFP, ChpT-eYFP, or CtrA-eYFP-14 (top row) and PAmCherry-PopZ (middle row) in a PopZ overexpression background; new poles marked with a white arrow. Scale bars: 2 µm. The percentages of total fluorescent signal at the new and old poles are indicated in blue, orange, and green for CckA, ChpT, and CtrA. Fluorescence intensity profiles shown along normalized cell length (n = 45, 52, 61 cells respectively). c. Overexpression of PopZ leads to a reduction in the expression of CtrA regulated genes. RT-qPCR of mRNA levels of five CtrA regulated genes at the 90-minute time point. Transcription was measured for wildtype cells (left) as well as cells overexpressing PopZ (right). Shown is the mean ± s.d.; n equals three biologically independent experiments repeated with similar results.
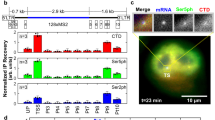
Extended Data Fig. 9 Spatially and temporally resolved transcriptional activity.
a. Time-resolved RT-qPCR of mRNA from an eyfp gene under the control of the CtrA-regulated P350 promoter integrated at four loci of the chromosome (L1, L3, L7, and L8, cf. Figure 3). Shown is mean ± s.d.; n = 3. Below plot: Replication times of these loci during the cell cycle, and cell cycle schematic of loci/dividing cells. Chromosome: black ovals. Red areas indicate polar PopZ microdomain, green tone indicates CtrA~P levels. Transcription is normalized to the five-minutes time point of L1. b. Time-resolved RT-qPCR of eyfp expressed from PxylX::eyfp at chromosomal loci L1 and L8 demonstrates a copy number effect. The observed increase at L1 and L8 represents the effect of chromosome replication, as xylose concentration and promotor activity are not expected to vary over time. Shown is mean ± s.d.; n = 3. c. eyfp transcription measured in cells at the 90 minute cell cycle time point at L1, L3, L7, and L8 in five genetic backgrounds. In the first three backgrounds, eyfp expression is controlled by the CtrA~P-regulated P350 promoter in the presence of: native wildtype ctrA (green), the phosphomimetic ctrA(D51E) driven by PxylX in a delta ctrA background (black), or ctrA driven by PxylX in a delta ctrA background (red). In the remaining backgrounds, eyfp expression is driven by the CtrA~P-regulated promoter PpilA in the presence of native wildtype ctrA (blue), and the xylose-driven promoter PxylX in the presence of xylose (gray). The wildtype ctrA, the phosphomimetic ctrA D51E, and eyfp driven by PxylX (green, black, and gray) are a subset of the data presented in Fig. 3b. The position-specific transcriptional output present for the CtrA~P-regulated P350 promoter in native conditions is lost when the sole CtrA copy is xylose-induced phosphomimetic CtrA(D51E) (black) but is recovered with xylose-inducible wildtype ctrA (red). Shown is mean + s.d.; n = 3. d. Simulated CtrA~P distribution profiles and their effect on transcription from P350. (left), possible CtrA~P profiles simulated with altered binding coefficient value between ChpT and PopZ at the new pole (cf. Extended Data Fig. 7a). Profile 4 (light brown) reflects wildtype parameters (Fig. 3a). (right) calculated transcript levels from P350 for each CtrA~P profile (Methods). Profile 4 was generated using wild-type parameters and best matched the experimental data (black, Fig. 3b).
Extended Data Fig. 10 High throughput assay to probe gene expression as a function of genomic position.
a. Graphical representation of the BacTRIP method. (I) A library of reporter plasmids generates a library of barcoded Caulobacter loci (purple, green, and red). Light gray: Tn5 mediated transposon. This includes a 5’ mosaic end (ME) followed by a SalI site, kanamycin resistance cassette (KanR), the origin of replication (ori), a promoter (gray), a gene of interest (yellow), a transcribed barcode (colors), a stop codon, the metK gene terminator from Caulobacter, and the 3’ ME. Transformation of the reporter plasmid library into Caulobacter introduces at most one barcode into each cell at a random locus (light brown box). Each barcoded strain reports local transcriptional activity at its locus. (II) DNA. Genomic DNA is used both to determine the integration locus (left) and the number of times each barcode was integrated (right). To determine the locus of each integrated barcode we digested the extracted DNA with SalI and used reverse PCR with mapping primers (Supplementary Table 8) to amplify fragments with an ori element for sequencing. To determine the number of times each barcode was integrated we amplified and sequenced the extracted DNA with counting primers (Supplementary Table 8). (III) RNA. We produced cDNA for sequencing from extracted, reverse-transcribed mRNA. The resulting quantity of inferred mRNA abundance was normalized by total copies of the barcode present in the library. b. BacTRIP results. We used three different CtrA binding sites to probe the wildtype CCNA_00350 promoter (PWT350), an engineered promoter in which we replaced the CtrA binding motif (red) of P350 with a consensus motif (PAT350), as well as a mutant site that is not recognized by CtrA (PMT350). We synchronized the three populations of cells and preformed BacTRIP on predivisional cells at the 90 minute time point. The bar graphs show abundance of 161, 156, and 148 barcoded eyfp as a function of locus distance from the new pole for pTripTn5- PWT350::eyfp, pTripTn5- PAT350::eyfp, and pTripTn5- PMT350::eyfp.
Supplementary information
Supplementary Information
Supplementary Notes 1–4, Supplementary Tables 1–8 and Supplementary References.
Supplementary Video 1
Single-molecule tracks of CtrA-eYFP-14. Exemplary raw data used to produce single-molecule trajectories of CtrA-eYFP-14. First track: entry to the new pole. (Data plotted in Fig. 1f, left). Second track: exit from the old pole. As noted in Methods, while not all single molecules used to produce tracks persisted this long before photobleaching, these results (reversible slowing of proteins within the poles) were consistently observed in 89 cells for CtrA-eYFP-14.
Source data
Source Data Fig. 1
Statistical Source Data for panels g and h.
Source Data Fig. 2
Statistical Source Data for panels b, d and e.
Source Data Fig. 3
Statistical Source Data for panel b.
Source Data Extended Data Fig. 1
SPR data for panel d and Statistical Source Data for panel e.
Source Data Extended Data Fig. 1
Full blots for panel b.
Source Data Extended Data Fig. 8
Representative RT-qPCR data for panel c.
Source Data Extended Data Fig. 8
Full blots for panel a.
Source Data Extended Data Fig. 9
Statistical Source Data for panels a–c.
Rights and permissions
About this article
Cite this article
Lasker, K., von Diezmann, L., Zhou, X. et al. Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nat Microbiol 5, 418–429 (2020). https://doi.org/10.1038/s41564-019-0647-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0647-7
This article is cited by
-
An experimental framework to assess biomolecular condensates in bacteria
Nature Communications (2024)
-
The molecular basis for cellular function of intrinsically disordered protein regions
Nature Reviews Molecular Cell Biology (2024)
-
Biomolecular condensates – extant relics or evolving microcompartments?
Communications Biology (2023)
-
Engineering synthetic biomolecular condensates
Nature Reviews Bioengineering (2023)
-
The material properties of a bacterial-derived biomolecular condensate tune biological function in natural and synthetic systems
Nature Communications (2022)