Abstract
Staphylococcus aureus is a major human pathogen that causes an array of infections ranging from minor skin infections to more serious infections, including osteomyelitis, endocarditis, necrotizing pneumonia and sepsis1. These more serious infections usually arise from an initial bloodstream infection and are frequently recalcitrant to antibiotic treatment1. Phagocytosis by macrophages and neutrophils is the primary mechanism through which S. aureus infection is controlled by the immune system2. Macrophages have been shown to be a major reservoir of S. aureus in vivo3, but the role of macrophages in the induction of antibiotic tolerance has not been explored. Here, we show that macrophages not only fail to efficiently kill phagocytosed S. aureus, but also induce tolerance to multiple antibiotics. Reactive oxygen species generated by respiratory burst attack iron–sulfur cluster-containing proteins, including TCA-cycle enzymes, result in decreased respiration, lower ATP and increased antibiotic tolerance. We further show that respiratory burst induces antibiotic tolerance in the spleen during a murine systemic infection. These results suggest that a major component of the innate immune response is antagonistic to the bactericidal activities of antibiotics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Additional data that support the findings of this study are available from the corresponding author upon request.
Change history
10 February 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41564-020-0679-z
References
Lowy, F. D. Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 (1998).
Rogers, D. E. Studies on bacteriemia. I. Mechanisms relating to the persistence of bacteriemia in rabbits following the intravenous injection of staphylococci. J. Exp. Med. 103, 713–742 (1956).
Surewaard, B. G. et al. Identification and treatment of the Staphylococcus aureus reservoir in vivo. J. Exp. Med. 213, 1141–1151 (2016).
Lehar, S. M. et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 527, 323–328 (2015).
Barcia-Macay, M., Seral, C., Mingeot-Leclercq, M. P., Tulkens, P. M. & Van Bambeke, F. Pharmacodynamic evaluation of the intracellular activities of antibiotics against Staphylococcus aureus in a model of THP-1 macrophages. Antimicrob. Agents Chemother. 50, 841–851 (2006).
Helaine, S. et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208 (2014).
Liu, Y. et al. Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo. J. Exp. Med. 213, 809–825 (2016).
Acocella, G., Carlone, N. A., Cuffini, A. M. & Cavallo, G. The penetration of rifampicin, pyrazinamide, and pyrazinoic acid into mouse macrophages. Am. Rev. Respir. Dis. 132, 1268–1273 (1985).
Pontes, M. H. & Groisman, E. A. Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci. Signal. 12, eaax3938 (2019).
Hobby, G. L. The antibacterial action of penicillin against Gram negative organisms. Science 100, 500–501 (1944).
Lopatkin, A. J. et al. Bacterial metabolic state more accurately predicts antibiotic lethality than growth rate. Nat. Microbiol. https://doi.org/10.1038/s41564-019-0536-0 (2019).
Conlon, B. P. et al. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 1, 16051 (2016).
Sonderholm, M. et al. The consequences of being in an infectious biofilm: microenvironmental conditions governing antibiotic tolerance. Int. J. Mol. Sci. 18, 2688 (2017).
Stewart, P. S. Antimicrobial tolerance in biofilms. Microbiol. Spectr. 3, MB-0010-2014 (2015).
Wang, Y. et al. Inactivation of TCA cycle enhances Staphylococcus aureus persister cell formation in stationary phase. Sci. Rep. 8, 10849 (2018).
Lobritz, M. A. et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl Acad. Sci. USA 112, 8173–8180 (2015).
Wood, T. K., Knabel, S. J. & Kwan, B. W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 79, 7116–7121 (2013).
Kwan, B. W., Valenta, J. A., Benedik, M. J. & Wood, T. K. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Ch. 57, 1468–1473 (2013).
Gardner, P. R. & Fridovich, I. Inactivation-reactivation of aconitase in Escherichia coli. A sensitive measure of superoxide radical. J. Biol. Chem. 267, 8757–8763 (1992).
Jang, S. & Imlay, J. A. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282, 929–937 (2007).
Castro, L., Rodriguez, M. & Radi, R. Aconitase is readily inactivated by peroxynitrite, but not by its precursor, nitric oxide. J. Biol. Chem. 269, 29409–29415 (1994).
Cohen, T. S. et al. S. aureus evades macrophage killing through NLRP3-dependent effects on mitochondrial trafficking. Cell Rep. 22, 2431–2441 (2018).
Abuaita, B. H., Schultz, T. L. & O’Riordan, M. X. Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe 24, 625–636 (2018).
Mitchell, J. B. et al. A low molecular weight antioxidant decreases weight and lowers tumor incidence. Free Radic. Biol. Med. 34, 93–102 (2003).
Jacobs, R. F. & Wilson, C. B. Activity of antibiotics in chronic granulomatous disease leukocytes. Pediatr. Res. 17, 916–919 (1983).
Sun, K. et al. Nox2-derived oxidative stress results in inefficacy of antibiotics against post-influenza S. aureus pneumonia. J. Exp. Med. 213, 1851–1864 (2016).
Kohanski, M. A., Dwyer, D. J., Hayete, B., Lawrence, C. A. & Collins, J. J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007).
Keren, I., Wu, Y., Inocencio, J., Mulcahy, L. R. & Lewis, K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339, 1213–1216 (2013).
Mosel, M., Li, L., Drlica, K. & Zhao, X. Superoxide-mediated protection of Escherichia coli from antimicrobials. Antimicrob. Agents Chemother. 57, 5755–5759 (2013).
Wu, Y., Vulic, M., Keren, I. & Lewis, K. Role of oxidative stress in persister tolerance. Antimicrob. Agents Chemother. 56, 4922–4926 (2012).
Wang, T., El Meouche, I. & Dunlop, M. J. Bacterial persistence induced by salicylate via reactive oxygen species. Sci. Rep. 7, 43839 (2017).
Hong, S. H., Wang, X., O’Connor, H. F., Benedik, M. J. & Wood, T. K. Bacterial persistence increases as environmental fitness decreases. Microb. Biotechnol. 5, 509–522 (2012).
Huang, X., Li, Y., Fu, M. & Xin, H. B. Polarizing macrophages in vitro. Methods Mol. Biol. 1784, 119–126 (2018).
Hall, J. D., Craven, R. R., Fuller, J. R., Pickles, R. J. & Kawula, T. H. Francisella tularensis replicates within alveolar type II epithelial cells in vitro and in vivo following inhalation. Infect. Immun. 75, 1034–1039 (2007).
Peloquin, C. A. et al. Population pharmacokinetic modeling of isoniazid, rifampin, and pyrazinamide. Antimicrob. Agents Chemother. 41, 2670–2679 (1997).
Szalek, E., Kaminska, A., Gozdzik-Spychalska, J., Grzeskowiak, E. & Batura-Gabryel, H. The PK/PD index (CMAX/MIC) for ciprofloxacin in patients with cystic fibrosis. Acta Pol. Pharm. 68, 777–783 (2011).
Nesseler, N. et al. High-dose continuous oxacillin infusion results in achievement of pharmacokinetics targets in critically ill patients with deep sternal wound infections following cardiac surgery. Antimicrob. Agents Chemother. 58, 5448–5455 (2014).
Barcia-Macay, M., Lemaire, S., Mingeot-Leclercq, M. P., Tulkens, P. M. & Van Bambeke, F. Evaluation of the extracellular and intracellular activities (human THP-1 macrophages) of telavancin versus vancomycin against methicillin-susceptible, methicillin-resistant, vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 58, 1177–1184 (2006).
Radlinski, L. et al. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol. 15, e2003981 (2017).
Ding, Y. et al. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc. Natl Acad. Sci.USA 111, E4981–E4990 (2014).
Cheung, A. L., Nast, C. C. & Bayer, A. S. Selective activation of sar promoters with the use of green fluorescent protein transcriptional fusions as the detection system in the rabbit endocarditis model. Infect. Immun. 66, 5988–5993 (1998).
Fey, P. D. et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4, e00537–e00512 (2013).
Acknowledgements
This work was supported primarily by NIH grant nos R01AI137273 and K22AI125501 to B.P.C. The Microscopy Services Laboratory is supported in part by the P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center. The oxygraph experiments were supported by grant no. R01CA211732 to Q.Z. The animal experiments were supported by grant nos R01AI137273 and K22AI125501 to B.P.C, and grant nos AI133236, AI139304, AI119073, AI136920 and the Yang Biomedical Scholars Award to E.A.M. The UNC Lineberger Animal Studies Core Facility, which is supported in part by an NCI Center Core support grant (grant no. CA16086) to the UNC Lineberger Comprehensive Cancer Center, assisted with the animal experiments. We are grateful to the Gastrointestinal Biology and Disease (CGIBD) Imaging and Histology Core, supported by NIH grant no. P30-DK 034987 awarded to the CGIBD Imaging and Histology Core, for processing the histology samples. We thank G. Jones and J. Arthur for their technical advice and equipment.
Author information
Authors and Affiliations
Contributions
B.P.C., S.E.R., N.J.W. and E.A.M. conceptualized the project. B.P.C. and S.E.R. wrote the manuscript. S.E.R., N.J.W., L.C.R., A.D.W., J.E.B. and L.L. performed the in vitro experiments. N.J.W. and J.E.B. performed the tissue culture experiments. S.E.R., N.J.W., L.C.R., J.E.B. and L.L. performed the animal experiments. Q.Z. provided equipment. S.E.R., N.J.W. and J.E.B. produced the figures. B.P.C. and E.A.M. provided funding for the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
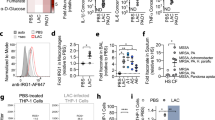
Extended Data Fig. 1 Induction of antibiotic tolerance by ROS.
(a) S. aureus strain COL or (b–e) HG003 was grown to mid-exponential phase in vitro and exposed to 80 μM menadione (MD), 0.125μg/ml mupirocin, acidified media (pH4.5), 5mM paraquat (PQ), 120 mM hydrogen peroxide (H2O2), 4mM N-acetyl cysteine (NAC) or 150mM thiourea (TU) for 20 min and either (a, c, d) challenged with 10µg/ml rifampicin or (b) plated for cfu or (e) OD600 was recorded every 30min for 10h. (a–d) At indicated times, an aliquot was removed and plated on tryptic soy agar (TSA) to enumerate cfu. Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (a, c) Student’s t-test (unpaired, two- tailed) or (d) One-Way ANOVA with Dunnett’s multiple comparison test.
Extended Data Fig. 2 ROS induces multidrug tolerance in S. aureus.
HG003 was grown to exponential phase in vitro and exposed to 80μM menadione (MD) for 20min prior to challenge with (a) 2.34μg/ml ciprofloxacin (cip), (b) 50μg/ml oxacillin (ox) or (c) 50μg/ml vancomycin (vanc). (d) Survival of S. aureus cells after internalization by unstimulated J774 macrophages (unstim.) or macrophages that were stimulated overnight with LPS/IFNγ (stim.). 40μg/ml cip was added at Time 0 (dotted lines). At the indicated times, an aliquot was removed, washed and plated to enumerate survivors. (e) % Survival of S. aureus cells treated with ciprofloxacin for 4h compared to the untreated control (data extrapolated from d). Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (a-c, e) the Student’s t-test (unpaired, two-tailed) or (d) One-Way ANOVA with Sidak’s multiple comparison test.
Extended Data Fig. 3 Growth cessation is insufficient to induce tolerance to rifampicin.
S. aureus strain HG003 was grown to mid-exponential phase in vitro and exposed to 80μM menadione (MD), 5mM paraquat (PQ), 120mM hydrogen peroxide (H2O2) or 30µg/ml chloramphenicol (Cam) for 20min and either (a) plated for cfu or (b) challenged with 10µg/ml rifampicin. At indicated times, an aliquot was removed and plated on tryptic soy agar (TSA) to enumerate cfu. Averages of n=3 biologically independent samples. Error bars represent standard deviation.
Extended Data Fig. 4 ROS decreases the metabolic activity of S. aureus.
HG003 was grown to mid-exponential phase and treated with 80μM menadione (MD), 5mM paraquat (PQ), 120mM hydrogen peroxide (H2O2), 4mM N-acetyl cysteine (NAC) or 5.5mM glucose (gluc) for (a–f, j) 2h or (g, h) 0.5h and then assayed for (a, e) succinate dehydrogenase (SDH) activity, (b, f) isocitrate dehydrogenase (IDH) activity and (d, g, j) aconitase activity. (c) Transcriptional activity of acn, sdh and icd genes was determined using promoter-gfp reporters. (h) ATP was quantified using a BactiterGlo Cell Viability Assay. (i) At indicated times, ROS production was measured using L-012 luminescent probe and expressed as a fold-increase compared to the untreated control. RLU denotes relative luminescence units. Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (a–c) Student’s t-test (unpaired, two-tailed) or (d, e, j) One-Way ANOVA with Dunnett’s multiple comparison test, (f–h) One-Way ANOVA with Sidak’s multiple comparison test or (i) Two-Way ANOVA with Sidak’s multiple comparison test.
Extended Data Fig. 5 Macrophage-derived ROS induce antibiotic tolerance in S. aureus.
(a, b) Survival of S. aureus strain HG003 after internalization by THP-1 macrophages. Where indicated, macrophages were pretreated with 20 μM BHA for 1 h prior to the addition of bacteria. (a) 10μg/ml rifampicin (rif) was added at Time 0 (dotted lines). (b) % rifampicin tolerant cells was determined by comparing survivors after 6h of rifampicin treatment to survivors of the corresponding untreated time point (extrapolated from a). (c) ROS production by THP-1 macrophages with or without BHA treatment was quantified using the luminescent probe L-012. RLU denotes relative luminescence units. (d, e) Survival of S. aureus strain HG003 (WT), HG003acnA::erm (acnA) and HG003sdhB::erm (sdhB) mutants after internalization by (d) unstimulated J774 macrophages or macrophages that were (e) pre-stimulated overnight with LPS/IFNγ. 10μg/ml rifampicin (rif) was added at Time 0 (dotted lines). Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using (a, d, e) One-Way ANOVA with Sidak’s multiple comparison test or (b-c) Student’s t-test (unpaired, two-tailed).
Extended Data Fig. 6 Host-derived ROS induces antibiotic tolerance in a mouse model of sepsis.
Sepsis was induced by intravenous injection of S. aureus strain HG003 into (a, b) C57BL/6J wild type (WT) and Ncf1−/− mutant mice or (c, d) wild type mice treated with or without 25mM tempol. At 24h.p.i., mice were treated daily with 25mg/kg rifampicin or the vehicle control. S. aureus cfu were enumerated from the kidney and liver at 48h.p.i. or 72h.p.i. The mean is indicated by a horizontal line and the limit of detection is indicated by the x-axis (a–d). (a, b) Wild type mice: vehicle/rifampicin n=9 animals, Ncf1-/- mice: vehicle n=10 / rifampicin n=11 animals. (c, d) control n=4 / tempol n=5 animals (at 24h p.i,) and control/tempol mice: vehicle/rifampicin n=10 animals (at 48h and 72h.p.i.). (e-f) Extracellular bacterial numbers were enumerated in the spleen and liver of control (n=3 animals) and tempol treated (n=4 animals) mice at 24h.p.i. prior to antibiotic challenge and error bars represent standard deviation. Statistical significance was determined using (a–d) Kruskal Wallis One-Way ANOVA with Dunn’s multiple comparison test or the (e, f) Mann- Whitney test.
Extended Data Fig. 7 Resistance does not contribute to menadione-induced antibiotic tolerance.
HG003 was grown to mid-exponential phase in vitro and exposed to 80μM menadione (MD) for 20min prior to addition of 10µg/ml rifampicin. At indicated times, an aliquot was removed and plated on tryptic soy agar (TSA) to enumerate survivors or TSA containing 10µg/ml rifampicin to enumerate rifampicin-resistant mutants (rifR). Averages of n=3 biologically independent samples. Error bars represent standard deviation. Statistical significance was determined using Student’s t-test (unpaired, two-tailed).
Supplementary information
Source data
Source Data Fig. 1
Raw data and statistical source data.
Source Data Fig. 2
Raw data and statistical source data.
Source Data Fig. 3
Raw data.
Source Data Fig. 4
Raw data and statistical source data.
Source Data Extended Data Fig. 1
Raw data and statistical source data.
Source Data Extended Data Fig. 2
Raw data and statistical source data.
Source Data Extended Data Fig. 3
Raw data and statistical source data.
Source Data Extended Data Fig. 4
Raw data and statistical source data.
Source Data Extended Data Fig. 5
Raw data and statistical source data.
Source Data Extended Data Fig. 6
Raw data and statistical source data.
Source Data Extended Data Fig. 7
Raw data and statistical source data.
Rights and permissions
About this article
Cite this article
Rowe, S.E., Wagner, N.J., Li, L. et al. Reactive oxygen species induce antibiotic tolerance during systemic Staphylococcus aureus infection. Nat Microbiol 5, 282–290 (2020). https://doi.org/10.1038/s41564-019-0627-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0627-y
This article is cited by
-
Comparative proteomic analysis of vancomycin-sensitive and vancomycin-intermediate resistant Staphylococcus aureus
European Journal of Clinical Microbiology & Infectious Diseases (2024)
-
Macrophage internalization creates a multidrug-tolerant fungal persister reservoir and facilitates the emergence of drug resistance
Nature Communications (2023)
-
Transcription tuned by S-nitrosylation underlies a mechanism for Staphylococcus aureus to circumvent vancomycin killing
Nature Communications (2023)
-
Roles of extracellular vesicles on macrophages in inflammatory bone diseases
Molecular and Cellular Biochemistry (2023)
-
Human serum triggers antibiotic tolerance in Staphylococcus aureus
Nature Communications (2022)