Abstract
The cytosolic appearance and propagation of bacteria cause overwhelming cellular stress responses that induce apoptosis under normal conditions. Therefore, successful bacterial colonization depends on the ability of intracellular pathogens to block apoptosis and to safeguard bacterial replicative niches. Here, we show that the cytosolic Gram-negative bacterium Shigella flexneri stalls apoptosis by inhibiting effector caspase activity. Our data identified lipopolysaccharide (LPS) as a bona fide effector caspase inhibitor that directly binds caspases by involving its O-antigen (O Ag) moiety. Bacterial strains that lacked the O Ag or failed to replicate within the cytosol were incapable of blocking apoptosis and exhibited reduced virulence in a murine model of bacterial infection. Our findings demonstrate how Shigella inhibits pro-apoptotic caspase activity, effectively delays coordinated host-cell demise and supports its intracellular propagation. Next to the recently discovered pro-inflammatory role of cytosolic LPS, our data reveal a distinct mode of LPS action that, through the disruption of the early coordinated non-lytic cell death response, ultimately supports the inflammatory breakdown of infected cells at later time points.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Jorgensen, I., Rayamajhi, M. & Miao, E. A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 17, 151–164 (2017).
Ashida, H., Kim, M. & Sasakawa, C. Manipulation of the host cell death pathway by Shigella. Cell. Microbiol. 16, 1757–1766 (2014).
Lamkanfi, M. & Dixit, V. M. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe 8, 44–54 (2010).
Tschopp, J., Thome, M., Hofmann, K. & Meinl, E. The fight of viruses against apoptosis. Curr. Opin. Genet. Dev. 8, 82–87 (1998).
Friedrich, A., Pechstein, J., Berens, C. & Luhrmann, A. Modulation of host cell apoptotic pathways by intracellular pathogens. Curr. Opin. Microbiol. 35, 88–99 (2017).
Danial, N. N. & Korsmeyer, S. J. Cell death: critical control points. Cell 116, 205–219 (2004).
Thornberry, N. A. & Lazebnik, Y. Caspases: enemies within. Science 281, 1312–1316 (1998).
Salvesen, G. S. & Ashkenazi, A. Snapshot: caspases. Cell 147, 476 (2011).
Bratton, S. B. & Salvesen, G. S. Regulation of the Apaf-1–caspase-9 apoptosome. J. Cell Sci. 123, 3209–3214 (2010).
Ashkenazi, A. & Dixit, V. M. Death receptors: signaling and modulation. Science 281, 1305–1308 (1998).
Ashkenazi, A. & Salvesen, G. Regulated cell death: signaling and mechanisms. Annu. Rev. Cell Dev. Biol. 30, 337–356 (2014).
Andree, M. et al. BID-dependent release of mitochondrial SMAC dampens XIAP-mediated immunity against Shigella. EMBO J. 33, 2171–2187 (2014).
Kashkar, H. et al. XIAP-mediated caspase inhibition in Hodgkin's lymphoma-derived B cells. J. Exp. Med. 198, 341–347 (2003).
Seeger, J. M. et al. The proteasome inhibitor bortezomib sensitizes melanoma cells toward adoptive CTL attack. Cancer Res. 70, 1825–1834 (2010).
Philpott, D. J., Yamaoka, S., Israel, A. & Sansonetti, P. J. Invasive Shigella flexneri activates NF-κB through a lipopolysaccharide-dependent innate intracellular response and leads to IL-8 expression in epithelial cells. J. Immunol. 165, 903–914 (2000).
Han, Z., Hendrickson, E. A., Bremner, T. A. & Wyche, J. H. A sequential two-step mechanism for the production of the mature p17:p12 form of caspase-3 in vitro. J. Biol. Chem. 272, 13432–13436 (1997).
Deveraux, Q. L. et al. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 17, 2215–2223 (1998).
Cummins, J. M. et al. X-linked inhibitor of apoptosis protein (XIAP) is a nonredundant modulator of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis in human cancer cells. Cancer Res. 64, 3006–3008 (2004).
Seeger, J. M. et al. Elevated XIAP expression alone does not confer chemoresistance. Br. J. Cancer 102, 1717–1723 (2010).
Kashkar, H. et al. XIAP targeting sensitizes Hodgkin lymphoma cells for cytolytic T-cell attack. Blood 108, 3434–3440 (2006).
Cossart, P. & Sansonetti, P. J. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304, 242–248 (2004).
Beuzon, C. R. et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19, 3235–3249 (2000).
Beutler, B. & Rietschel, E. T. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 3, 169–176 (2003).
Park, B. S. et al. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature 458, 1191–1195 (2009).
Das, S. et al. Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc. Natl Acad. Sci. USA 108, 2136–2141 (2011).
Hagar, J. A., Powell, D. A., Aachoui, Y., Ernst, R. K. & Miao, E. A. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341, 1250–1253 (2013).
Kayagaki, N. et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249 (2013).
Shi, J. et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014).
Kayagaki, N. et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015).
Kayagaki, N. et al. Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 (2011).
Raetz, C. R. & Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 (2002).
Kulp, A. & Kuehn, M. J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184 (2010).
Maurelli, A. T., Baudry, B., d'Hauteville, H., Hale, T. L. & Sansonetti, P. J. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect. Immun. 49, 164–171 (1985).
Sandlin, R. C. et al. Avirulence of rough mutants of Shigella flexneri: requirement of O antigen for correct unipolar localization of IcsA in the bacterial outer membrane. Infect. Immun. 63, 229–237 (1995).
Molinaro, A. et al. Full structural characterization of Shigella flexneri M90T serotype 5 wild-type R-LPS and its ΔgalU mutant: glycine residue location in the inner core of the lipopolysaccharide. Glycobiology 18, 260–269 (2008).
Kobayashi, T. et al. The Shigella OspC3 effector inhibits caspase-4, antagonizes inflammatory cell death, and promotes epithelial infection. Cell Host Microbe 13, 570–583 (2013).
Paciello, I. et al. Intracellular Shigella remodels its LPS to dampen the innate immune recognition and evade inflammasome activation. Proc. Natl Acad. Sci. USA 110, E4345–E4354 (2013).
Norris, M. H., Somprasong, N., Schweizer, H. P. & Tuanyok, A. Lipid A remodeling is a pathoadaptive mechanism that impacts lipopolysaccharide recognition and intracellular survival of Burkholderia pseudomallei. Infect. Immun. 86, e00360-18 (2018).
Martino, M. C. et al. Intravenous infection of virulent shigellae causes fulminant hepatitis in mice. Cell. Microbiol. 7, 115–127 (2005).
Kuida, K. et al. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature 384, 368–372 (1996).
Hong, M. & Payne, S. M. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol. Microbiol. 24, 779–791 (1997).
Ray, K., Marteyn, B., Sansonetti, P. J. & Tang, C. M. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. Microbiol. 7, 333–340 (2009).
Ashida, H. et al. Cell death and infection: a double-edged sword for host and pathogen survival. J. Cell Biol. 195, 931–942 (2011).
Hagar, J. A. & Miao, E. A. Detection of cytosolic bacteria by inflammatory caspases. Curr. Opin. Microbiol. 17, 61–66 (2014).
Yang, J., Zhao, Y. & Shao, F. Non-canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr. Opin. Immunol. 32, 78–83 (2015).
Clifton, D. R. et al. NF-κB-dependent inhibition of apoptosis is essential for host cell survival during Rickettsia rickettsii infection. Proc. Natl Acad. Sci. USA 95, 4646–4651 (1998).
Schwartz, J. T. et al. Francisella tularensis inhibits the intrinsic and extrinsic pathways to delay constitutive apoptosis and prolong human neutrophil lifespan. J. Immunol. 188, 3351–3363 (2012).
Mantis, N., Prevost, M. C. & Sansonetti, P. Analysis of epithelial cell stress response during infection by Shigella flexneri. Infect. Immun. 64, 2474–2482 (1996).
Lerouge, I. & Vanderleyden, J. O-antigen structural variation: mechanisms and possible roles in animal/plant–microbe interactions. FEMS Microbiol. Rev. 26, 17–47 (2002).
Jayakar, H. R. et al. A galU mutant of Francisella tularensis is attenuated for virulence in a murine pulmonary model of tularemia. BMC Microbiol. 11, 179 (2011).
Raynaud, C. et al. Role of the wbt locus of Francisella tularensis in lipopolysaccharide O-antigen biogenesis and pathogenicity. Infect. Immun. 75, 536–541 (2007).
Coutelle, O. et al. Embelin inhibits endothelial mitochondrial respiration and impairs neoangiogenesis during tumor growth and wound healing. EMBO Mol. Med. 6, 624–639 (2014).
Fortier, A. H. & Falk, L. A. Isolation of murine macrophages. Curr. Protoc. Immunol. 11, 14.1.1–14.1.9 (1994).
Salgado-Pabon, W. et al. Shigella impairs T lymphocyte dynamics in vivo. Proc. Natl Acad. Sci. USA 110, 4458–4463 (2013).
Utermohlen, O., Karow, U., Lohler, J. & Kronke, M. Severe impairment in early host defense against Listeria monocytogenes in mice deficient in acid sphingomyelinase. J. Immunol. 170, 2621–2628 (2003).
Nowak, J. et al. High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 72, 3277–3282 (2017).
Kashkar, H., Kronke, M. & Jurgensmeier, J. M. Defective Bax activation in Hodgkin B-cell lines confers resistance to staurosporine-induced apoptosis. Cell Death Differ. 9, 750–757 (2002).
Kashkar, H., Wiegmann, K., Yazdanpanah, B., Haubert, D. & Kronke, M. Acid sphingomyelinase is indispensable for UV light-induced Bax conformational change at the mitochondrial membrane. J. Biol. Chem. 280, 20804–20813 (2005).
Brinkmann, K. et al. Ubiquitin C-terminal hydrolase-L1 potentiates cancer chemosensitivity by stabilizing NOXA. Cell Rep. 3, 881–891 (2013).
Rezania, S. et al. Extraction, purification and characterization of lipopolysaccharide from Escherichia coli and Salmonella typhi. Avicenna J. Med. Biotechnol. 3, 3–9 (2011).
Westphal, O., Jann, K. & Himmelspach, K. Chemistry and immunochemistry of bacterial lipopolysaccharides as cell wall antigens and endotoxins. Prog. Allergy 33, 9–39 (1983).
Stennicke, H. R. & Salvesen, G. S. Caspases: preparation and characterization. Methods 17, 313–319 (1999).
Witt, A. et al. IAP antagonization promotes inflammatory destruction of vascular endothelium. EMBO Rep. 16, 719–727 (2015).
Acknowledgements
We thank P. Sansonetti (Institute Pasteur) for the Shigella strains M90T and BS176. We thank N. Robinson (University of Cologne) and D. Holden (Imperial College London) for the SalmonellaWT and SalmonellaΔSifA strains. We also thank O. Utermöhlen (University of Cologne) for the L. monocytogenes strain. We thank M. Menning, A. Manav, R. Hoppe, P. Glaser, T. Roth, D. Wagner-Stippich and K. Krönke-Wiegmann for their technical assistance. We thank the Imaging Facilities of CECAD and Collaborative Research Centre 670 (SFB670, Z2) and especially B. Martiny for preparing sections for electron microscopy. We thank B. Vogelstein (Johns Hopkins University) and M. Gyrd-Hansen (University of Oxford) for HCT116−/− XIAP cells. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) grant nos. CRC670, CRC1218 and CRU286.
Author information
Authors and Affiliations
Contributions
S.D.G. designed and carried out the majority of the experiments. M.F., J.M.S. and L.M.S. helped to perform viability assays, imaging and mice experiments. M.C. revised the paper. P.G.H. provided bacterial strains. S.H. supervised the SPR measurements. S.J.S. and G.S.S. provided recombinant caspase-3/-7. M.L.B. provided the ShigellaΔgalU strain. V.H., T.A.K. and M.K. provided essential reagents including cells and bacterial strains. H.K. designed the study. All authors analysed the data, discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Lack of caspase activity and apoptosis in cells infected with the invasive ShigellaM90T strain.
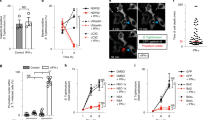
a, HeLa cells were either infected with ShigellaM90T (MOI: 30) for the indicated time points or exposed to 100 ng ml−1 Trail or 4 μM STS (3 h). Cytosolic extracts were isolated and analysed by Western blot with the indicated antibodies. PARP cleavage was examined in nuclear extracts. Tubulin served as a loading control. Viability was assessed using trypan blue exclusion assay (n = 3 biologically independent samples). b, Fluorescence microscopy of HeLa cells either infected with ShigellaM90T-dsRed (MOI: 30, 3 h) or exposed to 100 ng ml−1 Trail or 4 μM STS for 3 h and stained with Hoechst (blue). Scale bar, 20 μm. c, Analysis of intracellular bacterial burden by measuring CFU. HeLa cells were infected with the non-invasive ShigellaBS176 or the invasive ShigellaM90T strains and CFU was assessed in cellular lysates. Statistical significance to the corresponding non-infected value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 biologically independent samples). d, HCT cells were infected with the non-invasive ShigellaBS176 or the invasive ShigellaM90T strains. Cytosolic extracts of cells (MOI: 30, 3 h, 100 µg) were treated with recombinant active caspase-8 and caspase-3/-7 activity was measured. Statistical significance to the corresponding unstimulated value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 technical replicates of independent staining). e, HeLa cells were infected with increasing MOIs of ShigellaM90T and treated with 100 ng ml−1 Trail or 4 µM STS. Caspase-3/-7 activity (by Caspase-Glo 3/7 assay) was monitored after 3 h. Statistical significance to the corresponding non-infected value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 biologically independent samples). All experiments are representatives of at least two independent experiments and are presented as the mean ± s.d. **** p ≤ 0.0001; ** p ≤ 0.01; * p ≤ 0.05. Weights in Western blots are in kDa.
Extended Data Fig. 2 Shigella flexneri efficiently inhibits caspase-3/-7 enzymatic activity without involving the cellular caspase inhibitor XIAP.
a, Caco-2, HEK293T, HUVEC or HDMEC cells were infected with ShigellaM90T, cytosolic lysates were extracted and lysates were treated with active caspase-8. Caspase-3 proteolytic processing and activity was analysed. Statistical significance to the corresponding non-infected value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 technical replicates of independent staining). b, CFU analysis of intracellular ShigellaM90T in Caco-2, HEK293T, HUVEC and HDMEC cells (n = 3 biologically independent samples). c, Western blot analysis of whole cell lysates derived from shScr and shXIAP HeLa cells. Asterisk mark unspecific band. d, CFU analysis of intracellular ShigellaM90T in shScr and shXIAP HeLa cells. Statistical significance to the corresponding shScr value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 biologically independent samples). e, Western blot analysis of whole cell lysates derived from HCT WT and XIAP-/- cells. Asterisk mark unspecific band. f, CFU analysis of intracellular ShigellaM90T in HCT WT and HCT XIAP-/- cells. Statistical significance to the corresponding WT value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 biologically independent samples). g, Live cell imaging of shScr or shXIAP HeLa cells infected with ShigellaM90T and treated with 100 ng ml−1 Trail. Scale bar, 100 µm. All experiments are representatives of at least two independent experiments and are presented as the mean ± s.d. **** p ≤ 0.0001; ** p ≤ 0.01. Weights in Western blots are in kDa.
Extended Data Fig. 3 Cytosolic appearance of Gram-negative bacteria determines its capability to inhibit effector caspases.
a, CFU analysis of indicated intracellular bacteria. Statistical significance to the ShigellaM90T infected value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 biologically independent samples). b-c, Cytosolic extracts of HeLa cells infected with the indicated bacterial strains were activated with 100 ng ml−1 Trail or 4 µM STS and caspase-3/-7 activity was analysed. Statistical significance to the ShigellaM90T infected value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 technical replicates of independent staining). d, Live cell imaging of HeLa cells infected with the indicated pathogens and treated with 100 ng ml−1 Trail. Scale bar, 100 µm. e, HCT cells were infected with ShigellaM90T, Salmonella or SalmonellaΔSifA. Cytosolic extracts of cells (MOI: 30, 3 h, 100 µg) were incubated with recombinant active caspase-8 and caspase-3/-7 enzymatic activity were analysed. Statistical significance to the corresponding non-infected infected value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 technical replicates of independent staining). All experiments are representatives of at least two independent experiments and are presented as the mean ± s.d. **** p ≤ 0.0001; *** p ≤ 0.001; ** p ≤ 0.01; * p ≤ 0.05.
Extended Data Fig. 4 Cytosolic appearance of Gram-negative bacteria determines its capability to inhibit effector caspases.
a, HeLa cells were infected with Salmonella or SalmonellaΔSifA for 24 h and stained against endogenous Lamp-1 and Salmonella. Scale bar, 20 µm. b, Lamp-1 positive Salmonella at 3, 6 and 24 h p.i. (n = 15, 95, 38 for Salmonella at 3, 6, 24h, respectively. n = 72, 46, 61 for SalmonellaΔSifA at 3, 6, 24h, respectively of one representative experiment). c, Electron microscopy of HeLa cells infected with SalmonellaΔSifA at 3,6 and 8 h p.i. with a magnification of x10,000 (scale bar 1 µm), x25,000 (scale bar 500 nm), and x40,000 (scale bar 200 nm). Arrow point at phagocytic vacuole. d, CFU analysis of indicated intracellular Salmonella and SalmonellaΔSifA at 0, 3, 6 and 24 h p.i. Statistical significance to the Salmonella infected value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 biologically independent samples). e, HeLa cytosolic lysate was incubated with increasing concentrations of sonicated bacteria, activated with 1 U active recombinant caspase-8 and caspase-3 activity was monitored. Statistical significance to the corresponding untreated value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 technical replicates of independent staining). f, CFU analysis of the indicated bacteria before and after sonication to assess bacterial lysis. (n = 2 technical replicates). g, The indicated amounts of sonicated ShigellaM90T were treated with 20 mg ml−1 proteinase K and afterwards activated with recombinant active caspase-8. Caspase-3/-7 enzymatic activity was analysed. Statistical significance to the corresponding untreated value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 technical replicates of independent staining). All experiments are representatives of at least two independent experiments and are presented as the mean ± s.d. **** p ≤ 0.0001; *** p ≤ 0.001; ** p ≤ 0.01; * p ≤ 0.05.
Extended Data Fig. 5 LPS binds and inhibits effector caspases with its O-antigen region.
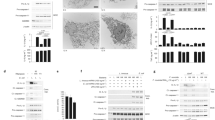
a, Recombinant procaspase-3 was incubated with LPS derived from Salmonella, ShigellaM90T, E. coliK12, Pseudomonas (P.a.) or E. coliO127:B8. Caspase-3 activity was measured after incubation with 1 U recombinant active caspase-8. Statistical significance to the corresponding untreated value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 technical replicates of independent staining). b, Recombinant procaspase-3 was incubated with LPS-biotin. LPS-biotin was then precipitated by streptavidin conjugated beads. Inputs and precipitates were analysed by Western blot. c, Cytosolic extracts derived from HeLa cells were incubated with increasing amounts of LPS-biotin alone or in conjunction with unlabelled lipid A or full-length LPS prior to precipitation by streptavidin conjugated beads. Inputs and precipitates were analysed by Western blot. Tubulin served as a loading control. d, Cytosolic extracts derived from HeLa cells (3x106 cells, 100 µg) were first incubated with increasing amounts of lipid A and then incubated with recombinant active caspase-8. Caspase-3 proteolytic processing and caspase-3/-7 enzymatic activity were analysed. Statistical significance to the corresponding untreated value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 technical replicates of independent staining). e, Coomassie gel of recombinant procaspase-3/-7 and their respective ∆prodomain mutants. f, SPR of binding capacity of catalytically inactive procaspase-3 and -7 or their Δprodomain mutants to E.coliO127:B8 and Pseudomonas aeruginosa (P.a.) LPS. Recombinant caspases were immobilised on a CM5 chip. Immobilised L-Lysine served as a control. g, Calculated representative dissociation constants (KD) of catalytically inactive procaspase-3 and -7 or their Δprodomain mutants to E. coliO127:B8 and P.a. LPS. h, SPR of binding capacity of catalytically inactive procaspase-3 and -7 or their Δprodomain mutants to Tri-DAP and lipid A. i, HeLa cells were electroporated with increasing concentrations of Ra LPS and caspase-3/-7 activity was measured after 16 h using Caspase 3/7 Glo. Statistical significance to the corresponding non-electroporated value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 9 biologically independent samples). All others NS. j, BMDMs were electroporated with 10 µg of the indicated LPS variants for 16 h and LDH release was analysed (n = 3 biologically independent samples). k, LPS concentration (in Endotoxin Units (EU) / ml) in 1 ng cytosolic extract was determined after 3 or 24 h p.i. Statistical significance to the corresponding ShigellaM90T-infected value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 2 technical replicates). All experiments are representatives of at least two independent experiments and are presented as the mean ± s.d. **** p ≤ 0.01; ** p ≤ 0.01; * p ≤ 0.05; ns > 0.05. Weights in Western blots are in kDa.
Extended Data Fig. 6 LPS-deficient ShigellaΔgalU fails to inhibit caspase activity and apoptosis.
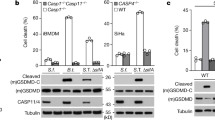
a, Optical density measurement at 600 nm (OD600) of ShigellaM90T or ShigellaΔgalU at the indicated time points to analyse growth defects. b, Electron microscopy of HeLa cells infected with ShigellaΔgalU. Scale bar, 1 µm (left panel), 200 nm (right panel). c, IL-8 release of HeLa cells infected with ShigellaM90T or ShigellaΔgalU. Statistical significance to the corresponding ShigellaM90T-infected value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 biologically independent samples). d, HeLa cells were infected with increasing MOIs of ShigellaΔgalU (upper panel) or ShigellaM90T (lower panel) treated with 100 ng ml−1 Trail or 4 µM STS for 6 h. Caspase-3 proteolytic processing was analysed by Western blot. Tubulin served as a loading control. e, HeLa cells were infected with ShigellaM90T or ShigellaΔgalU (MOI: 30, 6 h). Nuclei were stained with DAPI. Scale bar, 20 µm. f, CFU analysis of HeLa cells transfected with the indicated siRNAs (48-72 h; 100 nM) and infected with ShigellaM90T or ShigellaΔgalU (MOI: 30, 3 h). Statistical significance to the corresponding siScr value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 biologically independent samples). g, Live cell imaging of HCT cells, infected with ShigellaM90T or ShigellaΔgalU and treated with 100 ng ml−1 Trail (3 h). Scale bar, 20 µm. h, HCT cells were infected with ShigellaM90T or ShigellaΔgalU (MOI: 30, 3 h) and treated with the indicated concentrations of active caspase-8. Caspase-3 proteolytic processing and caspase-3/-7 enzymatic activity was analysed. Statistical significance to the corresponding untreated value was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 technical replicates of independent staining). i, CFU analysis of ShigellaΔgalU before and after sonication to assess bacterial lysis. Statistical significance was determined by two-tailed unpaired student’s t-test (n = 3 biologically independent samples). j, HeLa cells were electroporated with 0.1 µg of the indicated LPS variants for 16 h and LDH release was quantified. Statistical significance was determined by one-way ANOVA followed by Sidak’s post-analysis (n = 3 biologically independent samples). k, HCT cells were infected with ShigellaM90T or ShigellaΔgalU (MOI: 30, 3 h). Cellular lysates were isolated and analysed by Western blot with the indicated antibodies. Tubulin served as a loading control. l, Western Blot analysis of HeLa cytosolic lysates transfected with the indicated siRNAs (48-72 h; 100 nM) stained for the indicated antibodies (we were not able to detect caspase-5). Actin served as a loading control. All experiments are representatives of at least two independent experiments and are presented as the mean ± s.d. **** p ≤ 0.0001; *** p ≤ 0.001; ** p ≤ 0.01; * p ≤ 0.05. Weights in Western blots are in kDa.
Extended Data Fig. 7 Mice provoke efficient clearance and strong resistance against O-Ag-deficient Shigella strain.
a, Macroscopic images of ShigellaM90T or ShigellaΔgalU infected livers after 6 or 24 h p.i. b, Liver sections of mice infected for 24 h with ShigellaM90T or ShigellaΔgalU. Sections were stained against Shigella and DAPI. Scale bar, 2 mm. Detail scale bar, 500 µm. c, Confocal images of liver sections of ShigellaM90T or ShigellaΔgalU infected mice after 24 h p.i. Liver sections were stained with TUNEL, Shigella antibody and DAPI. Scale bar overview, 100 µm. Scale bar detail, 20 µm. d, Western blot analysis of whole cell lysates of BMDMs infected with ShigellaM90T or ShigellaΔgalU (MOI: 10, 3 h) stained for caspase-3 and -7. Tubulin served as a loading control. e, Western blot analysis of whole cell lysates of BMDM, liver or colon of caspase-3WT/WT and caspase-3-/- mice stained for caspase-3 and -7. Tubulin served as a loading control. f, Weight loss in percentage was calculated before infection and 48 h p.i. of caspase-3WT/WT and caspase-3-/- mice infected with ShigellaΔgalU. Statistical differences were analysed by two-tailed unpaired student’s t-test (n = 7 (Caspase-3WT/WT) and 9 (Caspase3-/-)). g, Histological analysis of livers from ShigellaΔgalU infected caspase-3WT/WT and caspase-3-/- mice. Sections were stained by H&E. Scale bar, 100 µm. All experiments are representatives of at least two independent experiments and are presented as mean ± s.d. * p ≤ 0.05. Weights in Western blots are in kDa.
Extended Data Fig. 8 Mice provoke efficient clearance and strong resistance against O-Ag-deficient Shigella strain.
a, Liver sections of ShigellaΔgalU infected caspase-3WT/WT and caspase-3-/- mice after 48 h p.i. Liver sections were stained with TUNEL, Shigella antibody and DAPI. Scale bar overview, 100 µm. Scale bar detail, 20 µm. b, TUNEL positive Shigella areas were counted in livers. 3-10 Shigella areas of each mouse were counted for each genotype and percentage of TUNEL positive areas was determined. Statistical differences were analysed by two-tailed unpaired student’s t-test (n = 40 (Caspase-3WT/WT) and 33 (Caspase3-/-) Shigella positive areas of 4 different mice). c, ShigellaM90T or ShigellaΔgalU were treated with complement preserved human or murine serum for 0, 2 and 4 h. OD600 (left panel) or CFU (right panel) was analysed (n = 3 technical replicates). Statistical significance to the corresponding untreated value was determined by one-way ANOVA followed by Sidak’s post-analysis. All others are ns (OD600: P = 0.5656; > 0.9999 for ShigellaM90T and ShigellaΔgalU to murine serum, respectively and CFU: P = 0.8806; 0.3889 for ShigellaM90T and ShigellaΔgalU to murine serum, respectively and ShigellaM90T to human serum P = 0.2996). All experiments are representatives of at least two independent experiments and are presented as mean ± s.d. **** p ≤ 0.0001; *** p ≤ 0.001; ** p ≤ 0.01.
Supplementary information
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 5
Statistical Source Data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 6
Statistical Source Data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical Source Data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 3
Statistical Source Data.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 6
Statistical Source Data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical Source Data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical Source Data.
Rights and permissions
About this article
Cite this article
Günther, S.D., Fritsch, M., Seeger, J.M. et al. Cytosolic Gram-negative bacteria prevent apoptosis by inhibition of effector caspases through lipopolysaccharide. Nat Microbiol 5, 354–367 (2020). https://doi.org/10.1038/s41564-019-0620-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0620-5
This article is cited by
-
Apoptosis and its pathways as targets for intracellular pathogens to persist in cells
Parasitology Research (2024)
-
Dying in self-defence: a comparative overview of immunogenic cell death signalling in animals and plants
Cell Death & Differentiation (2023)
-
Apoptotic cell death in disease—Current understanding of the NCCD 2023
Cell Death & Differentiation (2023)
-
Mitochondrial respiration controls neoangiogenesis during wound healing and tumour growth
Nature Communications (2020)