Abstract
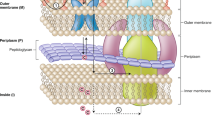
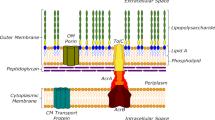
Gram-negative bacterial infections are a significant public health concern, and the lack of new drug classes for these pathogens is linked to the inability of most drug leads to accumulate inside Gram-negative bacteria1,2,3,4,5,6,7. Here, we report the development of a web application—eNTRyway—that predicts compound accumulation (in Escherichia coli) from its structure. In conjunction with structure–activity relationships and X-ray data, eNTRyway was utilized to re-design Debio-1452—a Gram-positive-only antibiotic8—into versions that accumulate in E. coli and possess antibacterial activity against high-priority Gram-negative pathogens. The lead compound Debio-1452-NH3 operates as an antibiotic via the same mechanism as Debio-1452, namely potent inhibition of the enoyl-acyl carrier protein reductase FabI, as validated by in vitro enzyme assays and the generation of bacterial isolates with spontaneous target mutations. Debio-1452-NH3 is well tolerated in vivo, reduces bacterial burden in mice and rescues mice from lethal infections with clinical isolates of Acinetobacter baumannii, Klebsiella pneumoniae and E. coli. This work provides tools for the facile discovery and development of high-accumulating compounds in E. coli, and a general blueprint for the conversion of Gram-positive-only compounds into broad-spectrum antibiotics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The main data supporting the findings of this study are available within the article and its Supplementary Information. Source data are available online for Figs. 2 and 3 and Extended Data Figs. 5–7 and 9. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.
Code availability
Source code for eNTRyway for local use is available on GitHub (https://github.com/HergenrotherLab/entry-cli).
References
Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 12, 371–387 (2013).
Cassini, A. et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66 (2019).
Theuretzbacher, U. et al. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet Infect. Dis. 19, e40–e50 (2019).
Tommasi, R., Brown, D. G., Walkup, G. K., Manchester, J. I. & Miller, A. A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 14, 529–542 (2015).
Tommasi, R., Iyer, R. & Miller, A. A. Antibacterial drug discovery: some assembly required. ACS Infect. Dis. 4, 686–695 (2018).
Silver, L. L. Topics in Medicinal Chemistry Vol. 25 (ed. Fisher, J. F. et al.) 31–67 (Springer, 2017).
Richter, M. F. & Hergenrother, P. J. The challenge of converting Gram-positive-only compounds into broad-spectrum antibiotics. Ann. NY Acad. Sci. 1435, 18–38 (2019).
Karlowsky, J. A., Kaplan, N., Hafkin, B., Hoban, D. J. & Zhanel, G. G. AFN-1252, a FabI inhibitor, demonstrates a Staphylococcus-specific spectrum of activity. Antimicrob. Agents Chemother. 53, 3544–3548 (2009).
Silver, L. L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 24, 71–109 (2011).
Rice, L. B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197, 1079–1081 (2008).
Ghosh, M. et al. Siderophore conjugates of daptomycin are potent inhibitors of carbapenem resistant strains of Acinetobacter baumannii. ACS Infect. Dis. 4, 1529–1535 (2018).
Li, X.-Z., Plésiat, P. & Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418 (2015).
Corbett, D. et al. Potentiation of antibiotic activity by a novel cationic peptide: potency and spectrum of activity of SPR741. Antimicrob. Agents Ch. 61, e00200–e00217 (2017).
Richter, M. F. et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 545, 299–304 (2017).
O’Boyle, N. M. et al. Open Babel: an open chemical toolbox. J. Cheminformatics 3, 33 (2011).
Oliphant, T. E. Guide to NumPy (Trelgol Publishing, 2006).
Seefeld, M. A. et al. Inhibitors of bacterial enoyl acyl carrier protein reductase (FabI): 2,9-disubstituted 1,2,3,4-tetrahydropyrido[3,4-b]indoles as potential antibacterial agents. Bioorg. Med. Chem. Lett. 11, 2241–2244 (2001).
Payne, D. J. et al. Discovery of a novel and potent class of FabI-directed antibacterial agents. Antimicrob. Agents Ch. 46, 3118–3124 (2002).
Yao, J. & Rock, C. O. Exogenous fatty acid metabolism in bacteria. Biochimie 141, 30–39 (2017).
Asturias, F. J. et al. Structure and molecular organization of mammalian fatty acid synthase. Nat. Struct. Mol. Biol. 12, 225–232 (2005).
McMurry, L. M., Oethinger, M. & Levy, S. B. Triclosan targets lipid synthesis. Nature 394, 531–532 (1998).
Rawat, R., Whitty, A. & Tonge, P. J. The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance. Proc. Natl Acad. Sci. USA 100, 13881–13886 (2003).
Zhu, L., Lin, J., Ma, J., Cronan, J. E. & Wang, H. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob. Agents Ch. 54, 689–698 (2010).
Parsons, J. B. & Rock, C. O. Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery? Curr. Opin. Microbiol. 14, 544–549 (2011).
Banevicius, M. A., Kaplan, N., Hafkin, B. & Nicolau, D. P. Pharmacokinetics, pharmacodynamics and efficacy of novel FabI inhibitor AFN-1252 against MSSA and MRSA in the murine thigh infection model. J. Chemother. 25, 26–31 (2013).
Hafkin, B., Kaplan, N. & Murphy, B. Efficacy and safety of AFN-1252, the first Staphylococcus-specific antibacterial agent, in the treatment of acute bacterial skin and skin structure infections, including those in patients with significant comorbidities. Antimicrob. Agents Ch. 60, 1695–1701 (2015).
Takhi, M. et al. Discovery of azetidine based ene-amides as potent bacterial enoyl ACP reductase (FabI) inhibitors. Eur. J. Med. Chem. 84, 382–394 (2014).
Ramnauth, J. et al. 2,3,4,5-Tetrahydro-1H-pyrido[2,3-b and e][1,4]diazepines as inhibitors of the bacterial enoyl ACP reductase, FabI. Bioorg. Med. Chem. Lett. 19, 5359–5362 (2009).
Sampson, P. B. et al. Spiro-naphthyridinone piperidines as inhibitors of S. aureus and E. coli enoyl-ACP reductase (FabI). Bioorg. Med. Chem. Lett. 19, 5355–5358 (2009).
Christie, S. M., Jinhong, R., Johnson, M. E. Enoyl reductase inhibitors with antibacterial activity. US patent 2,0180,072,666 A1 (2018).
Gerusz, V., Sonia, E., Oxoby, M., Denis, A. Novel heterocyclic acrylamides and their use as pharmaceuticals. US patent 8,846,711 B2 (2012).
Kaplan, N. et al. Mode of action, in vitro activity, and in vivo efficacy of AFN-1252, a selective antistaphylococcal FabI inhibitor. Antimicrob. Agents Ch. 56, 5865–5874 (2012).
Yao, J., Maxwell, J. B. & Rock, C. O. Resistance to AFN-1252 arises from missense mutations in Staphylococcus aureus enoyl-acyl carrier protein reductase (FabI). J. Biol. Chem. 288, 36261–36271 (2013).
Sivaraman, S., Zwahlen, J., Bell, A. F., Hedstrom, L. & Tonge, P. J. Structure–activity studies of the inhibition of FabI, the enoyl reductase from Escherichia coli, by triclosan: kinetic analysis of mutant FabIs. Biochemistry 42, 4406–4413 (2003).
Jackson, N., Czaplewski, L. & Piddock, L. J. V. Discovery and development of new antibacterial drugs: learning from experience?. J. Antimicrob. Chemoth. 73, 1452–1459 (2018).
Antibiotics currently in global clinical development PEW https://www.pewtrusts.org/en/research-and-analysis/data-visualizations/2014/antibiotics-currently-in-clinical-development (2019).
Lee, H. Y. et al. Reactive oxygen species synergize to potently and selectively induce cancer cell death. ACS Chem. Biol. 12, 1416–1424 (2017).
Palchaudhuri, R. et al. A small molecule that induces intrinsic pathway apoptosis with unparalleled speed. Cell Rep. 13, 2027–2036 (2015).
Llabani, E. et al. Diverse compounds from pleuromutilin lead to a thioredoxin inhibitor and inducer of ferroptosis. Nat. Chem. 11, 521–532 (2019).
Wayne, P. A. MO7: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically 11th edn (Clinical and Laboratory Standards Institute, 2018).
Acknowledgements
We are grateful to the NIH (AI136773 and GM118575) and University of Illinois for funding this work. We thank P. Tonge (State University of New York at Stony Brook) for the FabI expression vector, L. Dirikolu (School of Veterinary Medicine, Louisiana State University) for the pharmacokinetic data analysis and J. Cronan for helpful and insightful comments regarding this work.
Author information
Authors and Affiliations
Contributions
P.J.H., E.N.P. and B.S.D. conceived the study. E.N.P. performed the compound synthesis. B.S.D. constructed the eNTRyway web application. E.N.P., B.S.D. and E.J.G. performed the MIC experiments. B.S.D. performed the docking experiments, with the assistance of E.N.P. B.S.D. performed the enzyme inhibition assays and resistance studies. E.J.G. performed the accumulation assays. H.Y.L. performed the tolerability studies and pharmacokinetic study. G.W.L. conduced the efficacy studies. N.I. provided some of the clinical bacterial isolates. P.J.H. supervised this research. P.J.H., E.N.P and B.S.D. wrote the manuscript, with assistance from E.J.G.
Corresponding author
Ethics declarations
Competing interests
The University of Illinois has filed patents on some of the compounds described in this manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Overview of eNTRyway functionality and examples of visual outputs.
eNTRyway classifies molecules that are likely capable of accumulating in E. coli. eNTRyway calculates physiochemical properties of molecules and compares to a training set of compounds. SMILES strings for this analysis are generated in Open Babel using canonicalized atom order. Compounds containing an ionizable nitrogen (primary amines are most favorable), low three-dimensionality (globularity ≤ 0.25), and low flexibility (rotatable bonds ≤ 5) have a significantly higher probability of accumulating in E. coli. It is important to note that localized polarity and sterics about the amine are important considerations when implementing the eNTRy rules as these factors are not explicitly included in the eNTRYway results. Previously, it was found that increased amphiphilic moment (vsurf_A) correlated with increased accumulation while sterically encumbered primary amines displayed decreased accumulation14. Using eNTRyway to reanalyze the diverse compound collection published in Richter et al14 (Tables S2-S4; 188 compounds), we found that the accuracy is 84% (Supplementary Table 6).
Extended Data Fig. 2
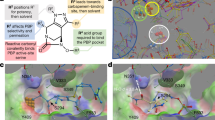
Docking of Debio-1452-NH3 in S. aureus FabI.
Extended Data Fig. 3 Synthesis of modified naphthyridinones.
Boc2O, di-tert-butyl dicarbonate; DIPEA, diisopropylamine; LHMDS, Lithium bis(trimethylsilyl)amide; THF, tetrahydrofuran.
Extended Data Fig. 4 Synthesis of amine- containing derivatives of Debio-1452.
Cy, cyclohexyl; DIPEA, diisopropylamine; DMA, dimethylacetamide.
Extended Data Fig. 5 Time-course accumulation of Debio- 1452 and derivatives in E. coli MG1655.
Bacterial cells were washed with and suspended in PBS. Cells were exposed to compound (50 µM final concentration) and incubated at 37 °C with shaking for 10 min, 30 min, 1 h, and 2 h. All cells were viable at these time points. Extra-cellular compound was removed, cells were lysed, and amount of compound in lysate was quantified by LC-MS/MS. Accumulation reported in nmol per 1012 colony-forming units (CFUs). Data shown represents the average of three independent experiments. Error bars represent the standard deviation of the mean. Measurements were compared by ordinary two-way ANOVA. Tukey’s multiple comparisons test was used to compare compounds at each timepoint. Statistical significance from Debio-1452 is indicated with asterisks (ns, not significant when p > 0.05 (Debio-1452-NH3, 10 min, p = 0.1719; Debio-1452-NH3, 0.5 h, p = 0.5522), *** p < 0.001, **** p < 0.0001).
Extended Data Fig. 6 Time-course accumulation of Debio- 1452 and derivatives in E. coli BW25113 and E. coli ΔtolC JW5503.
a. Debio-1452 accumulation over time. b. Debio-1452-NH3 accumulation over time. c. Compound 2 accumulation over time. d. Compound 3 accumulation over time. Bacterial cells were washed with and suspended in PBS. Cells were exposed to compound (50 µM final concentration) and incubated at 37 °C with shaking for 10 min, 30 min, 1 h, and 2 h. All cells were viable at these time points. Extra-cellular compound was removed, cells were lysed, and amount of compound in lysate was quantified by LC-MS/MS. Accumulation reported in nmol per 1012 colony-forming units (CFUs). Data shown represents the average of three independent experiments. Error bars represent standard deviation of the mean. Measurements were compared by ordinary two-way ANOVA. Sidak’s multiple comparisons test was used to compare compounds at each timepoint. Statistical significance of E. coli ΔtolC data points relative to E. coli BW25113 data points is indicated with asterisks (ns, not significant when p > 0.05 (b, 2.0 h, p = 0.0897; c, 10 min, p = 0.3571; c, 1.0 h, p = 0.9728; c, 2.0 h, p = 0.9894; d, 2.0 h, p = 0.0791), ** p < 0.01, *** p < 0.001, **** p < 0.0001).
Extended Data Fig. 7 Time-course accumulation of Debio- 1452 derivatives in P. aeruginosa PAO1.
Bacterial cells were washed with and suspended in PBS. Cells were exposed to compound (50 µM final) and incubated at 37 °C with shaking for 30 min, 1 h, and 2 h. All cells were viable at these time points. Extra-cellular compound was removed, cells were lysed, and amount of compound in lysate was quantified by LC-MS/MS. Accumulation reported in nmol per 1012 colony-forming units (CFUs). Data shown represents the average of three independent experiments. Error bars represent the standard deviation. Measurements were compared by ordinary two-way ANOVA. Tukey’s multiple comparisons test was used to compare compounds at each timepoint. Statistical significance from novobiocin, a negative antibiotic control, is indicated with asterisks (ns not significant when p > 0.05 (Debio-1452-NH3, 0.5 h, p = 0.3609, * p < 0.05, *** p < 0.001, **** p < 0.0001).
Extended Data Fig. 8 Location of mutated residues that confer resistance to Debio- 1452 and Debio-1452- NH3.
a. Structural alignment of E. coli FabI (periwinkle, PDB 4JQC) and S. aureus FabI (turquoise, PDB 4FS3). Residues mutated in E. coli are highlighted in red. Residues mutated in S. aureus are highlighted in blue. Debio-1452 from E. coli structure is green and from S. aureus is magenta. NADPH from S. aureus structure is pink. b. Same poses as a but without cartoon representation of protein backbone and NADPH. c. Distances from mutation sites and Debio-1452, either between centers of mass (COM) or the minimum pairwise distance between atoms within mutated residues and Debio-1452.
Extended Data Fig. 9 Pharmacokinetic analysis of Debio-1452- NH3.
C57BL/6 mice were treated with 50 mg/kg Debio-1452-NH3 via intraperitoneal injection, with three mice per time point (0, 15, 30, 45, 60, 120, 240 480, 960, and 1440 min). After the indicated time points, mice were sacrificed and the serum concentrations of Debio- 1452-NH3 were determined by LC-MS/MS. Data are shown as mean and represent the average of three independent experiments. The pharmacokinetic parameters shown in the figure above were calculated with a one-compartment model using a nonlinear regression program (Phoenix WinNonlin Version 8.1, Certara USA Inc., Princeton, NJ 08540 USA).
Extended Data Fig. 10 In vivo efficacy of Debio-1452-NH3 with strains that are less susceptible in cell culture.
a. Acute pneumonia infections initiated in CD-1 mice with A. baumannii M13100 (2.1x108 CFU/mouse, intranasal) were treated with vehicle (8 mice) or FabI inhibitor (8 mice per group) 6.5 and 23 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 48 h post-infection. b. Acute pneumonia infections initiated in CD-1 mice with K. pneumoniae S20595 (1.9x108 CFU/mouse, intranasal) were treated with vehicle (8 mice) or FabI inhibitor (8 mice per group) 6.5 and 22.5 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 48 h post-infection. c. Acute pneumonia infections initiated in CD-1 mice with K. pneumoniae BAA2473 (3.4x108 CFU/mouse, intranasal) were treated with vehicle (8 mice) or FabI inhibitor (8 mice per group) 6 and 22.5 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 48 h post-infection. d. Acute pneumonia infections initiated in CD-1 mice with K. pneumoniae AR0347 (2.5x108 CFU/mouse, intranasal) were treated with vehicle (8 mice) or FabI inhibitor (8 mice per group) 6 and 23 h post-infection (IV, 50 mg/kg), and the bacterial burden was evaluated 48 h post-infection. Drugs were formulated in 20% sulfobutyl ether(7) β-cyclodextrin from solid immediately before treatment. For the bacterial burden studies (a–d), data are shown as mean and error bars represent standard deviation. Significance was determined by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons. Statistical significance is indicated with asterisks (ns, not significant when p > 0.05 (a, p = 0.8582; b, p = 0.1019; c, p = 0.7600; d, p = 0.7536), **** p < 0.0001). Debio-1452-NH3 has an MIC of 16 µg/mL for all four strains used in these mouse models.
Supplementary information
Supplementary Information
Supplementary Tables 1–5, caption for Supplementary Table 6 and Supplementary Methods for the synthesis of compounds evaluated for biological activity.
Supplementary Table 6
Compound test set for calculation of eNTRyway accuracy.
Rights and permissions
About this article
Cite this article
Parker, E.N., Drown, B.S., Geddes, E.J. et al. Implementation of permeation rules leads to a FabI inhibitor with activity against Gram-negative pathogens. Nat Microbiol 5, 67–75 (2020). https://doi.org/10.1038/s41564-019-0604-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0604-5
This article is cited by
-
Alternative therapeutic strategies to treat antibiotic-resistant pathogens
Nature Reviews Microbiology (2024)
-
Structural and Biochemical Studies on Klebsiella Pneumoniae Enoyl-ACP Reductase (FabI) Suggest Flexible Substrate Binding Site
The Protein Journal (2024)
-
The small molecule raptinal can simultaneously induce apoptosis and inhibit PANX1 activity
Cell Death & Disease (2024)
-
Inhibiting fatty acid synthesis overcomes colistin resistance
Nature Microbiology (2023)
-
Tackling the outer membrane: facilitating compound entry into Gram-negative bacterial pathogens
npj Antimicrobials and Resistance (2023)