Abstract
Intestinal epithelial cells (IECs) act as a physical barrier separating the commensal-containing intestinal tract from the sterile interior. These cells have found a complex balance allowing them to be prepared for pathogen attacks while still tolerating the presence of bacterial or viral stimuli present in the lumen of the gut. Using primary human IECs, we probed the mechanisms that allow for such a tolerance. We discovered that viral infections emanating from the basolateral side of IECs elicit a stronger intrinsic immune response in comparison to lumenal apical infections. We determined that this asymmetric immune response is driven by the clathrin-sorting adaptor AP-1B, which mediates the polarized sorting of Toll-like receptor 3 (TLR3) towards the basolateral side of IECs. Mice and human IECs lacking AP-1B showed an exacerbated immune response following apical stimulation. Together, these results suggest a model where the cellular polarity program plays an integral role in the ability of IECs to partially tolerate apical commensals while remaining fully responsive to invasive basolateral pathogens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Peterson, L. W. & Artis, D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153 (2014).
Pott, J. & Hornef, M. Innate immune signalling at the intestinal epithelium in homeostasis and disease. EMBO Rep. 13, 684–698 (2012).
Fukata, M. & Arditi, M. The role of pattern recognition receptors in intestinal inflammation. Mucosal Immunol. 6, 451–463 (2013).
Arpaia, N. & Barton, G. M. Toll-like receptors: key players in antiviral immunity. Curr. Opin. Virol. 1, 447–454 (2011).
Barton, G. M. & Medzhitov, R. Toll-like receptor signaling pathways. Science 300, 1524–1525 (2003).
Odendall, C. & Kagan, J. C. Activation and pathogenic manipulation of the sensors of the innate immune system. Microbes Infect. 19, 229–237 (2017).
Chow, J., Franz, K. M. & Kagan, J. C. PRRs are watching you: localization of innate sensing and signaling regulators. Virology 479–480, 104–109 (2015).
Yu, S. & Gao, N. Compartmentalizing intestinal epithelial cell Toll-like receptors for immune surveillance. Cell. Mol. Life Sci. 72, 3343–3353 (2015).
Kagan, J. C. & Barton, G. M. Emerging principles governing signal transduction by pattern-recognition receptors. Cold Spring Harb. Persp. Biol. 7, a016253 (2014).
Rodriguez-Boulan, E. & Macara, I. G. Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15, 225–242 (2014).
Gewirtz, A. T., Navas, T. A., Lyons, S., Godowski, P. J. & Madara, J. L. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167, 1882–1885 (2001).
Lee, J. et al. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat. Cell Biol. 8, 1327–1336 (2006).
Price, A. E. et al. A map of Toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity 49, 560–575 (2018).
Madara, J. L., Stafford, J., Dharmsathaphorn, K. & Carlson, S. Structural analysis of a human intestinal epithelial cell line. Gastroenterology 92, 1133–1145 (1987).
Stanifer, M. L. et al. Reovirus intermediate subviral particles constitute a strategy to infect intestinal epithelial cells by exploiting TGF-β dependent pro-survival signaling. Cell. Microbiol. 18, 1831–1845 (2016).
Le Bivic, A., Real, F. X. & Rodriguez-Boulan, E. Vectorial targeting of apical and basolateral plasma membrane proteins in a human adenocarcinoma epithelial cell line. Proc. Natl Acad. Sci. USA 86, 9313–9317 (1989).
Folsch, H. Role of the epithelial cell-specific clathrin adaptor complex AP-1B in cell polarity. Cell. Logist. 5, e1074331 (2015).
Yamamoto, M. et al. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301, 640–643 (2003).
Takahashi, D. et al. The epithelia-specific membrane trafficking factor AP-1B controls gut immune homeostasis in mice. Gastroenterology 141, 621–632 (2011).
Cario, E. et al. Commensal-associated molecular patterns induce selective Toll-like receptor-trafficking from apical membrane to cytoplasmic compartments in polarized intestinal epithelium. Am. J. Pathol. 160, 165–173 (2002).
Hiemstra, I. H. et al. Intestinal epithelial cell transported TLR2 ligand stimulates Ly6C+ monocyte differentiation in a G-CSF dependent manner. Immunobiology 220, 1255–1265 (2015).
Pott, J. et al. Age-dependent TLR3 expression of the intestinal epithelium contributes to rotavirus susceptibility. PLoS Pathog. 8, e1002670 (2012).
Cario, E. & Podolsky, D. K. Differential alteration in intestinal epithelial cell expression of Toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect. Immun. 68, 7010–7017 (2000).
Mantegazza, A. R. et al. Adaptor protein-3 in dendritic cells facilitates phagosomal Toll-like receptor signaling and antigen presentation to CD4+ T cells. Immunity 36, 782–794 (2012).
Blasius, A. L. et al. Slc15a4, AP-3, and Hermansky–Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc. Natl Acad. Sci. USA 107, 19973–19978 (2010).
Setta-Kaffetzi, N. et al. AP1S3 mutations are associated with pustular psoriasis and impaired Toll-like receptor 3 trafficking. Am. J. Hum. Genet. 94, 790–797 (2014).
Ingle, H. et al. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-λ. Nat. Microbiol. 4, 1120–1128 (2019).
Heijmans, J. et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 3, 1128–1139 (2013).
Sturzenbecker, L. J., Nibert, M., Furlong, D. & Fields, B. N. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61, 2351–2361 (1987).
Broering, T. J., McCutcheon, A. M., Centonze, V. E. & Nibert, M. L. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 74, 5516–5524 (2000).
Shah, P. N. M. et al. Genome packaging of reovirus is mediated by the scaffolding property of the microtubule network. Cell. Microbiol. 19, e12765 (2017).
Boulant, S. et al. Similar uptake but different trafficking and escape routes of reovirus virions and infectious subvirion particles imaged in polarized Madin–Darby canine kidney cells. Mol. Biol. Cell 24, 1196–1207 (2013).
Kanaya, T. et al. Development of intestinal M cells and follicle-associated epithelium is regulated by TRAF6-mediated NF-κB signaling. J. Exp. Med. 215, 501–519 (2018).
Bartfeld, S. & Clevers, H. Organoids as model for infectious diseases: culture of human and murine stomach organoids and microinjection of Helicobacter pylori. J. Vis. Exp. 105, e53359 (2015).
Shi, W., Oshlack, A. & Smyth, G. K. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 38, e204 (2010).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 1544–6115 (2004).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Acknowledgements
This work was supported by a research grant from the Chica and Heinz Schaller Foundation and Deutsche Forschungsgemeinschaft (Project nos 240245660 (Project 14 of SFB 1129) and 278001972 (Project A09 of TRR186) to S.B.). This project received funding from the European Union’s Seventh Framework Programme under grant agreement no. 334336 (FP7-PEOPLE-2012-CIG). M.S. was supported by an Olympia Morata Fellowship from the Heidelberg University Hospital, the Brigitte-Schlieben Lange Program from the state of Baden Württemberg, Germany and the Dual Career Support from CellNetworks. M.M. is supported by the TRR186. K.P. and S.M. were supported by the SFB 1129. J.C.K. was supported by NIH grant no. 5R01AI093589.
Author information
Authors and Affiliations
Contributions
M.L.S. conceived and carried out experiments, interpreted the results and wrote the manuscript. M.M. performed and analysed the immunofluorescence imaging experiments. S.M., K.P. and D.A. assisted in collecting and isolating RNA. T.H. analysed the microarray data. T.K. and H.O. generated the knockout-mice organoids. C.O., V.H. and J.C.K. contributed to the concept of the study and critical discussions. S.Bartfeld designed and performed the microinjection experiments. S.Boulant conceived experiments, interpreted results and wrote the manuscript. The final version of the manuscript was approved by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Barrier function and polarized response by T84 cells.
T84 cells were seeded onto transwell inserts. (a). The rate of establishment of barrier function was measured by the trans- epithelial electrical resistance (TEER) using a chop stick electrode. TEER greater than 1000 Ohm/cm2 indicates a tight barrier and is marked with a dashed line. Each point represents the mean +/- sd. n=6. (b). Barrier integrity was assessed by dextran diffusion assay. T84 cells were allowed to grow on transwells and at the indicated time points 4kDa fluorescent FITC-Dextran was added to the apical chamber. Three hours post-incubation, basolateral media was collected and analysed by fluorometry to assess the amount of dextran which has diffused to the basolateral compartment of the transwells. Results represent the mean +/- sd. n=3. n.d.= not determined. (c) pBMCs were isolated and treated with 10-fold serial dilutions of the indicated TLR agonists (starting concentrations: TLR1/2 (PAM3CSK4, 1 μg/mL); TLR3 (1:1 HMW and LMW poly(I:C), 10 μg/mL); TLR4 (LPS-EK, 1 μg/mL); TLR5 (Flagellin-ST, 1 μg/mL); TLR7/8 (R848, 1 μg/mL), TLR9 (ODN 2395 5 μM)). Supernatants were harvested 24 h post-stimulation and were tested for IL-6 by ELISA. Results represent the mean +/- sd. n=3. (d) Polarized T84 cells were fed FITC-labelled poly(I:C) in a side specific manner. Cells were harvested at indicated time points, were washed with a quick acid wash to remove any non-internalized poly(I:C) bound to the cell surface. Following acid wash, cells were lysed and the relative amount of internalized FITC-labelled poly(I:C) was measured by spectrofluorometry. Results represent the mean +/- sd. n=3. P=values were calculated from a two-tailed unpaired t test using GraphPad Prizm.
Extended Data Fig. 2 Apical infection of T84 cells by MRV leads to more de novo virus production compared to basolateral infection.
Polarized T84 cells were infected apically or basolaterally with MRV. (a) Fluorescently labelled MRV was added to polarized T84 cells in a side specific manner. Cells were collected at indicated times points and were washed with a quick acid wash to remove any non-internalized virus particles bound to the cell surface. Following acid wash, cells were lysed, and the relative amount of internalized virus particles was measured by spectrofluorometry. Results show the mean +/- sd. n=3. (b) Infected cells were collected at the indicated time points and viral replication was assayed by q-RT-PCR against the MRV μ2 genome segment. Results are normalized to non-infected controls and show the mean +/- sd. n=3. (c) Same as b except virus replication was addressed by Western blot against the MRV non-structural protein μNS. Actin was used as a loading control (left panel). Representative figure is shown. The relative expression of μNS is normalized to actin is shown (right panel). Results shown the mean +/- sd. n=3. (d) Infected cells were fixed at indicated time points and immunofluorescence was performed against the non-structural protein μNS. The number of infected cells per field of view was counted. 10 fields of view were counted for each time point for each replicate. Results shown the mean +/- sd. n=3. (e) Polarized T84 cells were infected apically or basolaterally with MRV. Infected cells were collected at the indicated time points and viral replication was assayed by q- RT-PCR against the MRV μ2 genome segment. Results are normalized to non-infected controls and show the mean +/- sd. n=3. (a-e) P-values were calculated from a two-tailed unpaired t test using GraphPad Prizm.
Extended Data Fig. 3 Side specific induction of IFN production upon viral infection in the human intestinal epithelial lines.
(a-b) SKCO15 cells were seeded on transwells. Following establishment of barrier function, cells were infected apically or basolaterally with MRV. (a) 16 hpi, the production of type I (IFNβ) and type III (IFNλ2/3) IFNs was assessed by q-RT-PCR. Results are normalized to mock infected samples and show the mean +/- sd. n=3. (b) Efficiency of the primary MRV infection in SKCO15 cells was controlled by immunofluorescence against the non-structural protein μNS (red). Cell nuclei were stained using DAPI (blue). Three biological replicates were performed. Representative images are shown. Scale bar =100 μm. (c) T84 cells were infected apically or basolaterally with MRV. RNA samples were collected at the indicated times post-infection and production of type I (IFNβ) and type III (IFNλ2/3) interferons was measured by q-RT-PCR. Results are normalized to mock infected samples and show the mean +/- sd. n=3. (d) T84 cells were infected apically or basolaterally with MRV. Supernatants were collected at the indicated times post- infection and analysed by ELISA for the secretion of type III (IFNλ2/3) interferon. Results show the mean +/- sd. n=3. Black line indicates the limit of detection. (a-d) A=apical infection. B =basolateral infection. (a,d) P values were calculated using a two-tailed unpaired t test in GraphPad Prizm.
Extended Data Fig. 4 Stronger immune response observed during basolateral infection of hIECs is not due to differences in transcript stability.
(a) Polarized T84 cells were infected apically or basolaterally with MRV. 16 hpi cells were treated with actinomycin D to block transcription. RNA samples were collected at the indicated times post-actinomycin treatment. (b) Relative amounts of type I (upper panel) and type III IFN (lower panel) transcripts (normalized to the time of actinomycin treatment) were evaluated by q-RT-PCR (left panel). The half-life of each transcripts was determined (right panel). Results indicate the mean +/- sd. n=3. P values were determined from a two-tailed unpaired t test using GraphPad Prizm.
Extended Data Fig. 5 Basolateral infection of T84 cells by MRV leads to the upregulation of many immune related genes.
Polarized T84 cells were infected apically or basolaterally with MRV. 16 hpi, RNA was collected and subjected to microarray analysis. Expression values were quantile-normalized after background correction and log2-transformed. Unspecific filtering was performed by discarding genes with non- significant p-values in any sample. (a) Differentially expressed genes between apical infection and mock infected cells as well as between basolateral infection and mock infected cells were identified using the empirical Bayes approach and are shown in a Venn diagram. (b) Heatmap of expression values of significantly upregulated genes in apically and basolaterally infected cells were scaled across samples and hierarchical clustering was performed using euclidean distance and complete linkage. Gene values and their reference numbers can be found in the Supplementary Dataset. (c) KEGG analysis of significantly upregulated genes upon basolateral infection of T84 cells.
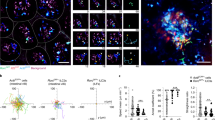
Extended Data Fig. 6 Basolateral infection of human organoids by MRV generates a stronger intrinsic immune response compared to apical infection.
(a) Schematic of the microinjection approach used to infect organoids specifically from either their apical or basolateral sides. (b-c) Human intestinal organoids were infected by MRV apically or basolaterally through microinjection. Virus infection was assessed 16 hpi for (b) the production of the non-structural viral protein μNS using Western blot analysis. Representative western blot is shown. and (c) the production of the MRV μ2 genome segment by q-RT-PCR. Results shown the mean +/- sd. n=3. (d) Generation of intrinsic immune response was assessed by quantifying the production of type I (IFNβ) and type III (IFNλ2/3) IFNs using q-RT-PCR. Results shown the mean +/- sd. n=3. (c-d) P values were calculated with a two-tailed unpaired t test using GraphPad Prizm. (e) The relative expression of TLR2-9 was evaluated in human intestinal organoids. Results are expressed as a ratio to the house keeping gene HPRT1. Results shown the mean +/- sd. n=3.
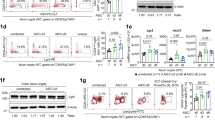
Extended Data Fig. 7 Knockdown of the clathrin adaptor AP-1B impacts the TLR3 mediated polarized immune response.
(a) Knock-down efficiency AP-1B was evaluated by q-RT-PCR. Results show the mean +/- sd. n=3. (b-d) The integrity of the barrier function of the AP-1B knock down T84 cells was evaluated by (b) indirect immunofluorescence of the tight junction protein Z-O1 (red). Representative image for immunofluorescence is shown. Scale bar =100 μm. (c) Dextran diffusion assay. Results show the mean +/- sd. n=3. and (d) TEER measurement. Results show the mean +/- sd. n=6. (e) Polarized WT and AP-1B knock-down T84 cells were infected with MRV in a side specific manner. Supernatants were collected at indicated time points and subjected to ELISA to monitor the amount of secreted type III IFNs. Results show the mean +/- sd. n=3. (f) AP-1B knock-down T84 cells were seeded on transwells and treated with indicated TLR agonists: TLR1/2 (PAM3CSK4, 1 μg/mL); TLR3 (1:1 HMW and LMW poly(I:C), 10 μg/mL); TLR4 (LPS-EK, 0.01 μg/mL); TLR5 (Flagellin-ST, 1 μg/mL); TLR7/8 (R848, 1 μg/mL), TLR9 (ODN 2395 5 μM). 6 h post-treatment RNA was harvested and evaluated for the upregulation of IL-6 by q-RT-PCR. Results are normalized to mock-treated samples and show the mean +/- s.d. n=3. (g) Same as f except type III IFN (IFNλ2/3) was evaluated. Results are normalized to mock-treated samples and show the mean +/- s.d. n=3. (e-g) P values were calculated from a two-tailed unpaired t test using GraphPad Prizm.
Extended Data Fig. 8 Inhibition of TRIF interferes with the polarized immune response generated by T84 cells.
(a-b) WT and AP-1B knock-down T84 cells were seeded on transwell inserts. (a) WT cells were treated with a TRIF inhibitor or a control peptide. T84 cells were infected with MRV in a side specific manner. RNA samples were harvested at indicated time points post-MRV infection and the intrinsic innate immune induction was analysed by q-RT-PCR for type III IFN (IFNλ2/3). Results show the mean +/- sd. n=4. (b) Same as a, except AP-1B knock-down cells were evaluated. Results show the mean +/- sd. n=4. (a-b) P values were calculated from a two-tailed unpaired t test using GraphPad Prizm. ns=not significant, * P < 0.05. ** P < 0.01. Statistics in panel b are between AP-1B and AP-1B+TRIFi. Exact p-values can be found in Supplementary Table 2.
Extended Data Fig. 9 TLR3 and AP-1B are not found together with immunofluorescence staining.
(a) TLR3 antibody specificity was controlled by transfecting HEK 293T (naturally non-TLR3 expressing) cells with a plasmid expressing TLR3. 48 h post-transfection cells were fixed and stained for TLR3 expression (red). Nuclei were stained with DAPI (blue). Representative images are shown. (b) T84 cells were seeded as spheroids. Five days post-seeding spheroids were fixed and stained for Laminin (red), and either TLR3 (green) or AP-1 (green). Nuclei were stained with DAPI (blue). Representative image is shown. Scale bar represents 20μm. (c) Same as b, except TLR3 and AP-1 were co-stained. Laminin (red), TLR3 (green), AP-1 (magenta), and nuclei were stained with DAPI (blue). Two representative images are shown. Scale bar represents 20μm in full size image and 5μm in zoom in.
Extended Data Fig. 10 The clathrin sorting adaptor protein AP-1 localizes TLR3 to the basolateral side of hIECs leading to an asymmetric IFN- mediated immune response.
Schematic showing TLR3 localization in polarized intestinal epithelial cells and its implication for side specific interferon induction upon viral infection.
Supplementary information
Supplementary Data
Microarray genes and values. Table shows expression values and gene reference numbers for microarray data shown in Extended Data 5.
Supplementary Tables
Tables 1 and 2 include the P values from Fig. 6 and Extended Data Fig. 8. Table 3 includes the primer sequences and accession numbers for the primers used in qPCR.
Source data
Source Data Fig. 2
Unprocessed western blot.
Source Data Extended Data Fig. 2
Unprocessed western blot.
Source Data Extended Data Fig. 6
Unprocessed western blot.
Rights and permissions
About this article
Cite this article
Stanifer, M.L., Mukenhirn, M., Muenchau, S. et al. Asymmetric distribution of TLR3 leads to a polarized immune response in human intestinal epithelial cells. Nat Microbiol 5, 181–191 (2020). https://doi.org/10.1038/s41564-019-0594-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0594-3
This article is cited by
-
Helicobacter pylori shows tropism to gastric differentiated pit cells dependent on urea chemotaxis
Nature Communications (2022)
-
Analogous comparison unravels heightened antiviral defense and boosted viral infection upon immunosuppression in bat organoids
Signal Transduction and Targeted Therapy (2022)
-
Organoids as host models for infection biology – a review of methods
Experimental & Molecular Medicine (2021)
-
Gastrointestinal epithelial innate immunity—regionalization and organoids as new model
Journal of Molecular Medicine (2021)