Abstract
To maintain prolonged infection of mammals, African trypanosomes have evolved remarkable surface coats and a system of antigenic variation1. Within these coats are receptors for macromolecular nutrients such as transferrin2,3. These must be accessible to their ligands but must not confer susceptibility to immunoglobulin-mediated attack. Trypanosomes have a wide host range and their receptors must also bind ligands from diverse species. To understand how these requirements are achieved, in the context of transferrin uptake, we determined the structure of a Trypanosoma brucei transferrin receptor in complex with human transferrin, showing how this heterodimeric receptor presents a large asymmetric ligand-binding platform. The trypanosome genome contains a family of around 14 transferrin receptors4, which has been proposed to allow binding to transferrin from different mammalian hosts5,6. However, we find that a single receptor can bind transferrin from a broad range of mammals, indicating that receptor variation is unlikely to be necessary for promiscuity of host infection. In contrast, polymorphic sites and N-linked glycans are preferentially found in exposed positions on the receptor surface, not contacting transferrin, suggesting that transferrin receptor diversification is driven by a need for antigenic variation in the receptor to prolong survival in a host.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
McCulloch, R. et al. Emerging challenges in understanding trypanosome antigenic variation. Emerg. Top. Life Sci. 1, 585–592 (2017).
Salmon, D. et al. A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell 78, 75–86 (1994).
Steverding, D. et al. ESAG 6 and 7 products of Trypanosoma brucei form a transferrin binding protein complex. Eur. J. Cell Biol. 64, 78–87 (1994).
Hertz-Fowler, C. et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS ONE 3, e3527 (2008).
Bitter, W., Gerrits, H., Kieft, R. & Borst, P. The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature 391, 499–502 (1998).
Gerrits, H., Mussmann, R., Bitter, W., Kieft, R. & Borst, P. The physiological significance of transferrin receptor variations in Trypanosoma brucei. Mol. Biochem. Parasitol. 119, 237–247 (2002).
Andrews, N. C. & Schmidt, P. J. Iron homeostasis. Annu. Rev. Physiol. 69, 69–85 (2007).
Luck, A. N. & Mason, A. B. Transferrin-mediated cellular iron delivery. Curr. Top. Membr. 69, 3–35 (2012).
Wally, J. et al. The crystal structure of iron-free human serum transferrin provides insight into inter-lobe communication and receptor binding. J. Biol. Chem. 281, 24934–24944 (2006).
Hall, D. R. et al. The crystal and molecular structures of diferric porcine and rabbit serum transferrins at resolutions of 2.15 and 2.60 Å, respectively. Acta Crystallogr. D 58, 70–80 (2002).
Congiu Castellano, A. et al. Structure–function relationship in the serotransferrin: the role of the pH on the conformational change and the metal ions release. Biochem. Biophys. Res. Commun. 198, 646–652 (1994).
Eckenroth, B. E., Steere, A. N., Chasteen, N. D., Everse, S. J. & Mason, A. B. How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH. Proc. Natl Acad. Sci. USA 108, 13089–13094 (2011).
Dautry-Varsat, A., Ciechanover, A. & Lodish, H. F. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc. Natl Acad. Sci. USA 80, 2258–2262 (1983).
Noinaj, N., Buchanan, S. K. & Cornelissen, C. N. The transferrin-iron import system from pathogenic Neisseria species. Mol. Microbiol. 86, 246–257 (2012).
Calmettes, C., Alcantara, J., Yu, R. H., Schryvers, A. B. & Moraes, T. F. The structural basis of transferrin sequestration by transferrin-binding protein B. Nat. Struct. Mol. Biol. 19, 358–360 (2012).
Steverding, D., Stierhof, Y. D., Fuchs, H., Tauber, R. & Overath, P. Transferrin-binding protein complex is the receptor for transferrin uptake in Trypanosoma brucei. J. Cell Biol. 131, 1173–1182 (1995).
Ligtenberg, M. J. et al. Reconstitution of a surface transferrin binding complex in insect form Trypanosoma brucei. EMBO J. 13, 2565–2573 (1994).
Salmon, D. et al. Characterization of the ligand-binding site of the transferrin receptor in Trypanosoma brucei demonstrates a structural relationship with the N-terminal domain of the variant surface glycoprotein. EMBO J. 16, 7272–7278 (1997).
Engstler, M. et al. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell 131, 505–515 (2007).
Steverding, D. The significance of transferrin receptor variation in Trypanosoma brucei. Trends Parasitol. 19, 125–127 (2003).
Cheng, Y., Zak, O., Aisen, P., Harrison, S. C. & Walz, T. Structure of the human transferrin receptor–transferrin complex. Cell 116, 565–576 (2004).
Williams, J. & Moreton, K. The distribution of iron between the metal-binding sites of transferrin human serum. Biochem. J. 185, 483–488 (1980).
Vanhollebeke, B. et al. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science 320, 677–681 (2008).
Mehlert, A., Wormald, M. R. & Ferguson, M. A. Modeling of the N-glycosylated transferrin receptor suggests how transferrin binding can occur within the surface coat of Trypanosoma brucei. PLoS Pathog. 8, e1002618 (2012).
Lane-Serff, H., MacGregor, P., Lowe, E. D., Carrington, M. & Higgins, M. K. Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor. eLife 3, e05553 (2014).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Wang, M. et al. “Anion clamp” allows flexible protein to impose coordination geometry on metal ions. Chem. Commun. (Camb.) 51, 7867–7870 (2015).
Blum, M. L. et al. A structural motif in the variant surface glycoproteins of Trypanosoma brucei. Nature 362, 603–609 (1993).
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D 60, 2210–2221 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Yang, N., Zhang, H., Wang, M., Hao, Q. & Sun, H. Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe. Sci. Rep. 2, 999 (2012).
Garman, E. F. & Grime, G. W. Elemental analysis of proteins by microPIXE. Prog. Biophys. Mol. Biol. 89, 173–205 (2005).
Grime, G. W., Dawson, M., Marsh, M., McArthur, I. C. & Watt, F. The Oxford submicron nuclear microscopy facility. Nucl. Instrum. Methods Phys. Res. B 54, 52–63 (1991).
Maxwell, J. A., Teesdale, W. J. & Campbell, J. L. The Guelph PIXE software package II. Nucl. Instrum. Methods Phys. Res. B 95, 407–421 (1995).
Grime, G. W. The “Q factor” method: quantitative microPIXE analysis using RBS normalisation. Nucl. Instrum. Methods Phys. Res. B 109–110, 170–174 (1996).
Acknowledgements
This work was supported by a research grant from the Medical Research Council (grant no. MR/R001138/1). C.E.T. holds a BBSRC iCASE studentship. M.K.H. is a Wellcome Investigator. The authors are grateful for the assistance of E. Lowe and the beamline scientists of beamline I03 at Diamond Light Source with crystallographic data collection. They also thank G. Grime at the Surrey Ion Beam Centre for assistance with μPIXE data collection, which was funded through EPSRC grant no. NS/A000059/1 to the UK National Ion Beam Centre.
Author information
Authors and Affiliations
Contributions
C.E.T. performed protein production, crystallization and surface plasmon resonance analysis. C.E.T. and M.K.H. determined the crystal structure. C.E.T., O.J.S.M. and M.C. conducted growth and expression analyses. P.G.W. and E.F.G. performed and analysed μPIXE experiments. A.L.G., S.R., T.J.V. and R.M. participated in design and experimental coordination. C.E.T., M.C. and M.K.H. devised the study and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
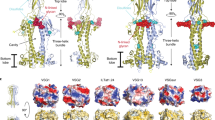
Extended Data Fig. 1 Phylogenic analysis of transferrin receptors.
a. Evolutionary trees showing the relatedness of different transferrin receptor variants within the blood stream expression sites of the Lister 427 strain. The BES1 and BES17 receptors are highlighted in red. b. Comparison of the BES17 ESAG6 and ESAG7 proteins. The black box indicates the signal sequence and the dashed black box indicates the GPI-anchor addition site. Blue cylinders represent the helical regions of the structure. Red stars mark residues which make direct contacts with human transferrin. Orange highlighting of the text indicates residues which differ between ESAG6 and ESAG7.
Extended Data Fig. 2 Data collection and refinement statistics.
Table of data collection and refinement statistics for crystal structures.
Extended Data Fig. 3 Comparison of the transferrin receptor with VSGs and the haptoglobin-haemoglobin receptor.
a. Structural alignment of the transferrin receptor with known VSG structures, 1VSG (orange), 2VSG (yellow) and 5LY9 (pink) and the Trypanosoma brucei haptoglobin–haemoglobin receptor (TbHpHbR, red). b. Sequence alignment of the two subunits of the transferrin receptor with the three VSGs. Orange highlighting indicates divergent residues.
Extended Data Fig. 4 Sequence alignment of mammalian transferrin variants analysed in this study.
a. Phylogenic tree of placental mammals adapted from ref. 36. b. Sequence alignment of selected transferrin variants. Variable residues are highlighted in orange and residues which directly contact the BES17 transferrin receptor are indicated by red stars. Green circles identify residues which contact the human transferrin receptor.
Extended Data Fig. 5 Table of interactions.
Table of interactions between the trypanosome transferrin receptor and transferrin.
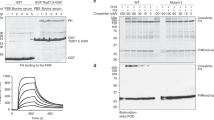
Extended Data Fig. 6 Surface plasmon resonance analysis.
Measurement of kinetic binding parameters for transferrin receptor variants to transferrins. BES1mut has a G139R mutation in ESAG6 and S246Y I229V and C223R in ESAG7. ESAG6 is e6 and ESAG7 is e7 in the table.
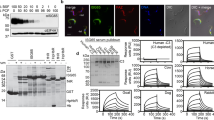
Extended Data Fig. 7 Elemental analysis by mPIXE to determine the number of Fe ions per transferrin or transferrin:TfR complex at different pH values.
Plots of counts per channel for emission induced by different photon energies, annotated with the element responsible for emission. Comparison of the quantities of Fe and S, together with knowledge of the protein sequences, and the numbers of S atoms, allowed determination of the number of Fe ions per transferrin or transferrin:TfR complex. Data shown is representative of three technical replicates.
Extended Data Fig. 8 Comparison of the sequences of BES17 and BES1 transferrin receptors.
Sequence alignments of ESAG6 subunits and ESAG7 subunits. Numbering is according to the BES17 sequence, which matches that of the crystal structure. Residues highlighted in orange vary between the BES1 and BES17 variants. Residues indicated by a red star directly contact human transferrin in the BES17 receptor. Residues indicated by a blue triangle are those mutated in the BES1mut mutant. Residues boxed with a continuous line represent the putative signal peptides while those boxed with a discontinuous line represent the GPI-anchor addition sequence.
Extended Data Fig. 9 SPR analysis of transferrin receptor variants and mutants.
a. Analysis of the binding of chimeric receptors formed between the ESAG6 and ESA7 subunits of the BES1 and BES17 transferrin receptor variants to human transferrin by surface plasmon resonance. b. Analysis of the binding to human and rat transferrin of mutants of the BES1 transferrin receptor. Each concentration series was performed once.
Extended Data Fig. 10 Analysis of transferrin receptor expression during growth in sera from different mammals.
a. Sequences of ESAG7 from different transferrin receptor variants within the blood stream expression sites of the Lister 427 strain, showing nucleotide sequences equivalent to 778 to 837 in the BES1 sequence. Below this are shown the predominant sequences found in Trypanosoma brucei cells grown in serum exclusively containing cow, horse, rabbit and pig transferrin for 309 hours. The red lettering and red star indicate a sequence difference at position 798 in which a T is unique to the BES1 receptor. b. Sequencing chromatograms of the expressed transferrin receptor variants in cells, showing nucleotides equivalent to 791 to 805 in the BES1 sequence. The star marks residue 798, which is T in BES1 and C in the other receptors in the Lister 427 strain. The top chromatogram shows that Trypanosoma brucei grown in fetal calf serum (cow) for 72 hours predominantly express BES1. These cells were transferred into media containing serum from cow, pig, horse or rabbit and were grown for a further 119 hours (the 191h time point) and 309 hours (the 381h time point) and the chromatograms show the transferrin receptor sequences expressed at these times remains predominantly BES1, with lower levels of other receptors in all four sera. A similar outcome was seen for ESAG6 sequences (not shown).
Supplementary information
Supplementary Table 1
Sequences used for sequence entropy calculations.
Rights and permissions
About this article
Cite this article
Trevor, C.E., Gonzalez-Munoz, A.L., Macleod, O.J.S. et al. Structure of the trypanosome transferrin receptor reveals mechanisms of ligand recognition and immune evasion. Nat Microbiol 4, 2074–2081 (2019). https://doi.org/10.1038/s41564-019-0589-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0589-0
This article is cited by
-
Cryo-EM structures of Trypanosoma brucei gambiense ISG65 with human complement C3 and C3b and their roles in alternative pathway restriction
Nature Communications (2023)
-
A multifaceted strategy to improve recombinant expression and structural characterisation of a Trypanosoma invariant surface protein
Scientific Reports (2022)
-
Invariant surface glycoprotein 65 of Trypanosoma brucei is a complement C3 receptor
Nature Communications (2022)