Abstract
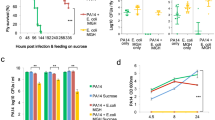
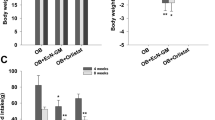
The microbiota confers colonization resistance, which blocks Salmonella gut colonization1. As diet affects microbiota composition, we studied whether food composition shifts enhance susceptibility to infection. Shifting mice to diets with reduced fibre or elevated fat content for 24 h boosted Salmonella Typhimurium or Escherichia coli gut colonization and plasmid transfer. Here, we studied the effect of dietary fat. Colonization resistance was restored within 48 h of return to maintenance diet. Salmonella gut colonization was also boosted by two oral doses of oleic acid or bile salts. These pathogen blooms required Salmonella’s AcrAB/TolC-dependent bile resistance. Our data indicate that fat-elicited bile promoted Salmonella gut colonization. Both E. coli and Salmonella show much higher bile resistance than the microbiota. Correspondingly, competitive E. coli can be protective in the fat-challenged gut. Diet shifts and fat-elicited bile promote S. Typhimurium gut infections in mice lacking E. coli in their microbiota. This mouse model may be useful for studying pathogen–microbiota–host interactions, the protective effect of E. coli, to analyse the spread of resistance plasmids and assess the impact of food components on the infection process.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
16S rRNA raw reads have been deposited at the European Nucleotide Archive (ENA) with accession no. PRJEB33890. All other data needed to evaluate the conclusions in this Article are presented in the paper or the Supplementary Information. Any additional data can be requested from the corresponding author.
References
Stecher, B., Berry, D. & Loy, A. Colonization resistance and microbial ecophysiology: using gnotobiotic mouse models and single-cell technology to explore the intestinal jungle. FEMS Microbiol. Rev. 37, 793–829 (2013).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Desai, M. S. et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353 (2016).
Barthel, M. et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858 (2003).
Brugiroux, S. et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2, 16215 (2016).
Stecher, B. et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl Acad. Sci. USA 109, 1269–1274 (2012).
Arkan, M. C. et al. IKK-β links inflammation to obesity-induced insulin resistance. Nat. Med. 11, 191–198 (2005).
Kim, K. A., Gu, W., Lee, I. A., Joh, E. H. & Kim, D. H. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS ONE 7, e47713 (2012).
Moya-Perez, A., Neef, A. & Sanz, Y. Bifidobacterium pseudocatenulatum CECT 7765 reduces obesity-associated inflammation by restoring the lymphocyte-macrophage balance and gut microbiota structure in high-fat diet-fed mice. PLoS ONE 10, e0126976 (2015).
Hwang, D. H., Kim, J. A. & Lee, J. Y. Mechanisms for the activation of Toll-like receptor 2/4 by saturated fatty acids and inhibition by docosahexaenoic acid. Eur. J. Pharm. 785, 24–35 (2016).
Stecher, B., Maier, L. & Hardt, W. D. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 11, 277–284 (2013).
Reddy, B. S., Mangat, S., Sheinfil, A., Weisburger, J. H. & Wynder, E. L. Effect of type and amount of dietary fat and 1,2-dimethylhydrazine on biliary bile acids, fecal bile acids and neutral sterols in rats. Cancer Res. 37, 2132–2137 (1977).
Reddy, B. S. Diet and excretion of bile acids. Cancer Res. 41, 3766–3768 (1981).
Cummings, J. H. et al. Influence of diets high and low in animal fat on bowel habit, gastrointestinal transit time, fecal microflora, bile acid and fat excretion. J. Clin. Invest. 61, 953–963 (1978).
Begley, M., Gahan, C. G. & Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651 (2005).
Islam, K. B. et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141, 1773–1781 (2011).
Urdaneta, V. & Casadesus, J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 4, 163 (2017).
MacConkey, A. T. Bile salt media and their advantages in some bacteriological examinations. J. Hyg. 8, 322–334 (1908).
Buckley, A. M. et al. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 8, 847–856 (2006).
Nishino, K., Latifi, T. & Groisman, E. A. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 126–141 (2006).
Devkota, S. et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487, 104–108 (2012).
Stecher, B. et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007).
Winter, S. E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010).
Winter, S. E. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013).
Raffatellu, M. et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486 (2009).
Litvak, Y., Byndloss, M. X. & Baumler, A. J. Colonocyte metabolism shapes the gut microbiota. Science 362, eaat9076 (2018).
Miki, T., Goto, R., Fujimoto, M., Okada, N. & Hardt, W. D. The bactericidal lectin RegIIIβ prolongs gut colonization and enteropathy in the streptomycin mouse model for Salmonella diarrhea. Cell Host Microbe 21, 195–207 (2017).
Miki, T., Holst, O. & Hardt, W. D. The bactericidal activity of the C-type lectin RegIIIβ against Gram-negative bacteria involves binding to lipid A. J. Biol. Chem. 287, 34844–34855 (2012).
Mukherjee, S. et al. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature 505, 103–107 (2014).
Lopez, C. A. et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3, e00143-12 (2012).
Berg, R. D. The indigenous gastrointestinal microflora. Trends Microbiol. 4, 430–435 (1996).
Piddock, L. J. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4, 629–636 (2006).
Wotzka, S. Y. et al. Microbiota stability in healthy individuals after single-dose lactulose challenge—a randomized controlled study. PLoS ONE 13, e0206214 (2018).
Litvak, Y. et al. Commensal Enterobacteriaceae protect against Salmonella colonization through oxygen competition. Cell Host Microbe 25, 128–139 (2019).
Sana, T. G. et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl Acad. Sci. USA 113, E5044–E5051 (2016).
Deriu, E. et al. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37 (2013).
Nedialkova, L. P. et al. Temperate phages promote colicin-dependent fitness of Salmonella enterica serovar Typhimurium. Environ. Microbiol. 18, 1591–1603 (2016).
Conway, T. & Cohen, P. S. Commensal and pathogenic Escherichia coli metabolism in the gut. Microbiol. Spectr. 3, MBP-0006-2014 (2015).
Baucheron, S. et al. AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium DT104. Antimicrob. Agents Chemother. 48, 3729–3735 (2004).
Eaves, D. J., Ricci, V. & Piddock, L. J. Expression of acrB, acrF, acrD, marA and soxS in Salmonella enterica serovar Typhimurium: role in multiple antibiotic resistance. Antimicrob. Agents Chemother. 48, 1145–1150 (2004).
Urdaneta, V. & Casadesus, J. Adaptation of Salmonella enterica to bile: essential role of AcrAB-mediated efflux. Environ. Microbiol. 20, 1405–1418 (2018).
Theriot, C. M. et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5, 3114 (2014).
Rozwandowicz, M. et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137 (2018).
Hoiseth, S. K. & Stocker, B. A. Aromatic-dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 (1981).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Grant, A. J. et al. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol. 6, e74 (2008).
Gil, D. & Bouche, J. P. ColE1-type vectors with fully repressible replication. Gene 105, 17–22 (1991).
Kaiser, P. et al. Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol. 12, e1001793 (2014).
Moor, K. et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 544, 498–502 (2017).
Sturm, A. et al. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7, e1002143 (2011).
Li, H. et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 6, 8292 (2015).
Apprill, A., McNally, S., Parsons, R. & Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecol. 75, 129–137 (2015).
Parada, A. E., Needham, D. M. & Fuhrman, J. A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016).
Dey, N. et al. Regulators of gut motility revealed by a gnotobiotic model of diet–microbiome interactions related to travel. Cell 163, 95–107 (2015).
Fuhrer, T., Heer, D., Begemann, B. & Zamboni, N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 83, 7074–7080 (2011).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Yin, S. et al. Factors affecting separation and detection of bile acids by liquid chromatography coupled with mass spectrometry in negative mode. Anal. Bioanal. Chem. 409, 5533–5545 (2017).
Wiegand, I., Hilpert, K. & Hancock, R. E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008).
Lambert, R. J. & Pearson, J. Susceptibility testing: accurate and reproducible minimum inhibitory concentration (MIC) and non-inhibitory concentration (NIC) values. J. Appl. Microbiol. 88, 784–790 (2000).
Cremer, J., Arnoldini, M. & Hwa, T. Effect of water flow and chemical environment on microbiota growth and composition in the human colon. Proc. Natl Acad. Sci. USA 114, 6438–6443 (2017).
Vandeputte, D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017).
Soetaert, K., Petzoldt, T. & Setzer, R. W. Solving differential equations in R: Package deSolve. J. Stat. Softw. 33, 1–25 (2010).
R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2014); https://www.R-project.org/
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
Acknowledgements
We thank members of the Hardt laboratory and R. Stocker for helpful scientific discussions and the RCHCI staff (especially K. Holzinger, D. Mollenhauer and S. Nowok) for excellent support of our animal work. E.S. is supported by the Swiss National Science Foundation (SNF, Marie Heim-Vögtlin award PMPDP3_158364 and Ambizione award PZ00P3_136742) and the Gebert Rüf ‘Microbials’ programme (GRS-073/17). W.-D.H. is supported by the SNF (310030_153074 and 310030B_173338/1; Sinergia CRSII_154414/1; NRP 72 407240-167121), ETH Zurich (ETH-33 12-2), the Novartis Freenovation Programme and the Promedica Foundation. S.S. is supported by ETH Zurich and the Helmut Horten Foundation. E.B. is supported by a Boehringer Ingelheim Fonds PhD Stipend and E.G. by a grant from the Monique Dornonville de la Cour Foundation. B.S. is supported by the German Research Foundation (STE 1971/4-2 SPP 1656/2) and the German Center for Infection Research (DZIF).
Author information
Authors and Affiliations
Contributions
S.Y.W. contributed to Figs. 1–3 and Supplementary Figs. 2–5, 7, 8, 10, 11, 16, 18, 22 and 23, B.N. to Supplementary Fig. 24, L.M. and A.T. to Fig. 2e and Supplementary Fig. 12, A.O.B. and J.P. to Fig. 2a, M.A. to Fig. 3a,b and Supplementary Figs. 13–15, D.L.B. to Supplementary Figs. 19 and 21b and Supplementary Tables 4 and 5, M. Zünd and S.S. to Supplementary Figs. 7 and 26, A.H. to Supplementary Fig. 21, E.B. and M.D. to Fig. 1e and Supplementary Fig. 6, K.M. to Fig. 1b–d, M.K. to Fig. 4 and Supplementary Figs. 4, 17, 18g and 25, D.H. and E.S. to Fig. 2d and Supplementary Fig. 23, M.B. and B.S. to Supplementary Fig. 7b, T.D. to Supplementary Fig. 19, M. Zimmermann, T.F. and U.S. to Supplementary Figs. 9 and 18a–b,f and E.G. to Supplementary Fig. 20. A.J.M and B.S. (Oligo mice) performed the experiments and analysed the data. S.Y.W., M.K., B.N., L.M., D.H., E.S. and W.-D.H. designed the experiments. S.Y.W., M.K. and W.-D.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Discussion, Supplementary Tables 1–6, Supplementary Figs. 1–26 and Supplementary References.
Supplementary Data 1
Mass spectrometry data for Supplementary Fig. 9.
Rights and permissions
About this article
Cite this article
Wotzka, S.Y., Kreuzer, M., Maier, L. et al. Escherichia coli limits Salmonella Typhimurium infections after diet shifts and fat-mediated microbiota perturbation in mice. Nat Microbiol 4, 2164–2174 (2019). https://doi.org/10.1038/s41564-019-0568-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0568-5
This article is cited by
-
Assessing microbiome population dynamics using wild-type isogenic standardized hybrid (WISH)-tags
Nature Microbiology (2024)
-
High-throughput anaerobic screening for identifying compounds acting against gut bacteria in monocultures or communities
Nature Protocols (2024)
-
Impact of enteric bacterial infections at and beyond the epithelial barrier
Nature Reviews Microbiology (2023)
-
Short-term dietary changes can result in mucosal and systemic immune depression
Nature Immunology (2023)
-
Impact of horizontal gene transfer on emergence and stability of cooperative virulence in Salmonella Typhimurium
Nature Communications (2022)