Abstract
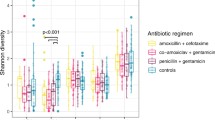
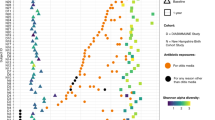
Hospitalized preterm infants receive frequent and often prolonged exposures to antibiotics because they are vulnerable to infection. It is not known whether the short-term effects of antibiotics on the preterm infant gut microbiota and resistome persist after discharge from neonatal intensive care units. Here, we use complementary metagenomic, culture-based and machine learning techniques to study the gut microbiota and resistome of antibiotic-exposed preterm infants during and after hospitalization, and we compare these readouts to antibiotic-naive healthy infants sampled synchronously. We find a persistently enriched gastrointestinal antibiotic resistome, prolonged carriage of multidrug-resistant Enterobacteriaceae and distinct antibiotic-driven patterns of microbiota and resistome assembly in extremely preterm infants that received early-life antibiotics. The collateral damage of early-life antibiotic treatment and hospitalization in preterm infants is long lasting. We urge the development of strategies to reduce these consequences in highly vulnerable neonatal populations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Assembled functional metagenomic contigs, shotgun metagenomic reads, shotgun genomic reads and assemblies have been deposited to NCBI GenBank and SRA under BioProject ID PRJNA489090.
Code availability
The software packages used in this study are free and open source. Analysis scripts used here (and associated usage notes) are available from the authors on reasonable request.
References
Sommer, F. & Bäckhed, F. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238 (2013).
Pantoja-Feliciano, I. G. et al. Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 7, 1112–1115 (2013).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Cox, L. M. et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721 (2014).
Abrahamsson, T. R. et al. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 44, 842–850 (2014).
Livanos, A. E. et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol. 1, 16140 (2016).
Cho, I. et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626 (2012).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Trasande, L. et al. Infant antibiotic exposures and early-life body mass. Int. J. Obes. 37, 16–23 (2013).
Hviid, A., Svanstrom, H. & Frisch, M. Antibiotic use and inflammatory bowel diseases in childhood. Gut 60, 49–54 (2011).
Penders, J. et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA birth cohort study. Gut 56, 661–667 (2007).
Arrieta, M.-C. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7, 307ra152 (2015).
Ahmadizar, F. et al. Early life antibiotic use and the risk of asthma and asthma exacerbations in children. Pediatr. Allergy Immunol. 28, 430–437 (2017).
Stokholm, J. et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 9, 141 (2018).
Missaghi, B., Barkema, H., Madsen, K. & Ghosh, S. Perturbation of the human microbiome as a contributor to inflammatory bowel disease. Pathogens 3, 510–527 (2014).
Tremlett, H. et al. Gut microbiota in early pediatric multiple sclerosis: a case−control study. Eur. J. Neurol. 23, 1308–1321 (2016).
Russell, S. L. et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447 (2012).
Zanvit, P. et al. Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nat. Commun. 6, 8424 (2015).
Azad, M. B., Bridgman, S. L., Becker, A. B. & Kozyrskyj, A. L. Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int. J. Obes. 38, 1290–1298 (2014).
Boursi, B., Mamtani, R., Haynes, K. & Yang, Y.-X. The effect of past antibiotic exposure on diabetes risk. Eur. J. Endocrinol. 172, 639–648 (2015).
Shaw, S. Y., Blanchard, J. F. & Bernstein, C. N. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am. J. Gastroenterol. 105, 2687–2692 (2010).
Ungaro, R. et al. Antibiotics associated with increased risk of new-onset Crohn’s disease but not ulcerative colitis: a meta-analysis. Am. J. Gastroenterol. 109, 1728–1738 (2014).
Kronman, M. P., Zaoutis, T. E., Haynes, K., Feng, R. & Coffin, S. E. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics 130, e794–e803 (2012).
Lexmond, W. S. et al. Involvement of the iNKT cell pathway is associated with early-onset eosinophilic esophagitis and response to allergen avoidance therapy. Am. J. Gastroenterol. 109, 646–657 (2014).
Blencowe, H. et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet 379, 2162–2172 (2012).
Liu, L. et al. Global, regional, and national causes of under-5 mortality in 2000–15: an updated systematic analysis with implications for the sustainable development goals. Lancet 388, 3027–3035 (2016).
Stoll, B. J. Neurodevelopmental and growth impairment among extremely low-birth-weight infants with neonatal infection. JAMA 292, 2357 (2004).
Flannery, D. D. et al. Temporal trends and center variation in early antibiotic use among premature infants. JAMA Netw. Open 1, e180164 (2018).
Rose, G. et al. Antibiotic resistance potential of the healthy preterm infant gut microbiome. PeerJ 5, e2928 (2017).
Rahman, S. F., Olm, M. R., Morowitz, M. J. & Banfield, J. F. Machine learning leveraging genomes from metagenomes identifies influential antibiotic resistance genes in the infant gut microbiome. mSystems 3, e00123-17 (2018).
Pärnänen, K. et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 9, 3891 (2018).
Hourigan, S. K. et al. Comparison of infant gut and skin microbiota, resistome and virulome between neonatal intensive care unit (NICU) environments. Front. Microbiol. 9, 1361 (2018).
Gibson, M. K. et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat. Microbiol. 1, 16024 (2016).
Fouhy, F. et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob. Agents Chemother. 56, 5811–5820 (2012).
Greenwood, C. et al. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of enterobacter. J. Pediatr. 165, 23–29 (2014).
Stewart, C. J. et al. Preterm gut microbiota and metabolome following discharge from intensive care. Sci. Rep. 5, 17141 (2015).
Zwittink, R. D. et al. Association between duration of intravenous antibiotic administration and early-life microbiota development in late-preterm infants. Eur. J. Clin. Microbiol. Infect. Dis. 37, 475–483 (2018).
Moles, L. et al. Preterm infant gut colonization in the neonatal ICU and complete restoration 2 years later. Clin. Microbiol. Infect. 21, 936–936 (2015).
The American College of Obstetricians and Gynecologists Committee on Obstetric Practice Society for Maternal-Fetal Medicine Committee opinion No 579: Definition of term pregnancy. Obstet. Gynecol. 122, 1139–1140 (2013).
Raju, T. N. K., Higgins, R. D., Stark, A. R. & Leveno, K. J. Optimizing care and outcome for late-preterm (near-term) infants: a summary of the workshop sponsored by the national institute of child health and human development. Pediatrics 118, 1207–1214 (2006).
Truong, D. T. et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903 (2015).
Franzosa, E. A. et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968 (2018).
Bradley, P. H. & Pollard, K. S. Proteobacteria explain significant functional variability in the human gut microbiome. Microbiome 5, 36 (2017).
Lindberg, T. P. et al. Preterm infant gut microbial patterns related to the development of necrotizing enterocolitis. J. Matern. Fetal Neonatal Med. 18, 1–10 (2018).
Warner, B. B. et al. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet 387, 1928–1936 (2016).
Abrahamsson, T. R. et al. Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 129, 434–440 (2012).
Kriss, M., Hazleton, K. Z., Nusbacher, N. M., Martin, C. G. & Lozupone, C. A. Low diversity gut microbiota dysbiosis: drivers, functional implications and recovery. Curr. Opin. Microbiol. 44, 34–40 (2018).
Subramanian, S. et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421 (2014).
Forsberg, K. J. et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111 (2012).
Hsieh, E. et al. Medication use in the neonatal intensive care unit. Am. J. Perinatol. 31, 811–822 (2013).
Jia, B. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573 (2017).
Crofts, T. S., Gasparrini, A. J. & Dantas, G. Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 15, 422–434 (2017).
Wyres, K. L. & Holt, K. E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 45, 131–139 (2018).
Navon-Venezia, S., Kondratyeva, K. & Carattoli, A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275 (2017).
Goldstone, R. J. & Smith, D. G. E. A population genomics approach to exploiting the accessory ‘resistome’ of Escherichia coli. Microb. Genom. 3, e000108 (2017).
Kaminski, J. et al. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput. Biol. 11, e1004557 (2015).
Breiman, L. Random Forests. Mach. Learn. 45, 5–32 (2001).
Carl, M. A. et al. Sepsis from the gut: the enteric habitat of bacteria that cause late-onset neonatal bloodstream infections. Clin. Infect. Dis. 58, 1211–1218 (2014).
Zhang, X., Feng, Y., Zhou, W., McNally, A. & Zong, Z. Cryptic transmission of ST405 Escherichia coli carrying bla NDM-4 in hospital. Sci. Rep. 8, 390 (2018).
Izdebski, R. et al. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J. Antimicrob. Chemother. 70, 48–56 (2015).
Gurnee, E. A. et al. Gut colonization of healthy children and their mothers with pathogenic ciprofloxacin-resistant Escherichia coli. J. Infect. Dis. 212, 1862–1868 (2015).
Kim, H. B. et al. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 3582–3584 (2009).
Fevre, C., Passet, V., Weill, F.-X., Grimont, P. A. D. & Brisse, S. Variants of the Klebsiella pneumoniae OKP Chromosomal beta-lactamase are divided into two main groups, OKP-A and OKP-B. Antimicrob. Agents Chemother. 49, 5149–5152 (2005).
Gasparrini, A. J. et al. Antibiotic perturbation of the preterm infant gut microbiome and resistome. Gut Microbes 7, 443–449 (2016).
Furtado, I. et al. Enterococcus faecium and Enterococcus faecalis in blood of newborns with suspected nosocomial infection. Rev. Inst. Med. Trop. Sao Paulo 56, 77–80 (2014).
Akturk, H. et al. Vancomycin resistant Enterococci colonization in a neonatal intensive care unit: who will be infected? J. Matern. Fetal Neonatal Med. 29, 3478–3482 (2016).
Brooks, B. et al. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome 2, 1 (2014).
Fernández-Canigia, L., Cejas, D., Gutkind, G. & Radice, M. Detection and genetic characterization of β-lactamases in Prevotella intermedia and Prevotella nigrescens isolated from oral cavity infections and peritonsillar abscesses. Anaerobe 33, 8–13 (2015).
Singh, B. et al. Probiotics for preterm infants: a national retrospective cohort study. J. Perinatol. 39, 533–539 (2019).
Kerr-Wilson, C. O., Mackay, D. F., Smith, G. C. S. & Pell, J. P. Meta-analysis of the association between preterm delivery and intelligence. J. Publ. Health 34, 209–216 (2012).
Johnson, S. et al. Academic attainment and special educational needs in extremely preterm children at 11 years of age: the EPICure study. Arch. Dis. Child. Fetal Neonatal Ed. 94, F283–F289 (2009).
Bhutta, A. T., Cleves, M. A., Casey, P. H., Cradock, M. M. & Anand, K. J. S. Cognitive and behavioral outcomes of school-aged children who were born preterm: a meta-analysis. JAMA 288, 728–737 (2002).
Tinnion, R., Gillone, J., Cheetham, T. & Embleton, N. Preterm birth and subsequent insulin sensitivity: a systematic review. Arch. Dis. Child. 99, 362–368 (2014).
Parkinson, J. R. C., Hyde, M. J., Gale, C., Santhakumaran, S. & Modi, N. Preterm birth and the metabolic syndrome in adult life: a systematic review and meta-analysis. Pediatrics 131, e1240–e1263 (2013).
Crump, C., Winkleby, M. A., Sundquist, K. & Sundquist, J. Risk of hypertension among young adults who were born preterm: a Swedish national study of 636,000 births. Am. J. Epidemiol. 173, 797–803 (2011).
Kowalski, R. R. et al. Elevated blood pressure with reduced left ventricular and aortic dimensions in adolescents born extremely preterm. J. Pediatr. 172, 75–80 (2016).
Crump, C., Winkleby, M. A., Sundquist, J. & Sundquist, K. Risk of asthma in young adults who were born preterm: a Swedish national cohort study. Pediatrics 127, e913–e920 (2011).
Lum, S. et al. Nature and severity of lung function abnormalities in extremely pre-term children at 11 years of age. Eur. Respir. J. 37, 1199–1207 (2011).
La Rosa, P. S. et al. Patterned progression of bacterial populations in the premature infant gut. Proc. Natl Acad. Sci. USA 111, 12522–12527 (2014).
Planer, J. D. et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534, 263–266 (2016).
Baym, M. et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS ONE 10, e0128036 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Zhu, W., Lomsadze, A. & Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38, e132 (2010).
Gibson, M. K., Forsberg, K. J. & Dantas, G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 9, 207–216 (2015).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277 (2000).
Schmieder, R. & Edwards, R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 6, e17288 (2011).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Ondov, B. D. et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132 (2016).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Mohamed, J. A., Huang, W., Nallapareddy, S. R., Teng, F. & Murray, B. E. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by enterococcus faecalis. Infect. Immun. 72, 3658–3663 (2004).
Acknowledgements
This work is supported in part by awards to G.D. through the National Institute of General Medical Sciences of the National Institutes of Health (R01 GM099538), the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI123394) and the US Centers for Disease Control and Prevention (200-2016-91955); to P.I.T. through the National Institutes of Health (5P30 DK052574 (Biobank, DDRCC)); to G.D., P.I.T. and B.B.W. through the Eunice Kennedy Shriver National Institute Of Child Health and Human Development of the National Institutes of Health (R01 HD092414); to P.I.T. and B.B.W. through the Children’s Discovery Institute at St Louis Children’s Hospital and Washington University School of Medicine; and to A.J.G. through a NIGMS training grant award number T32 GM007067 (J. Skeath, principal investigator) and from the NIDDK Pediatric Gastroenterology Research Training Program award number T32 DK077653 (P.I.T., principal investigator). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. We thank members of the Dantas laboratory for helpful discussion of the manuscript, and staff from the Edison Family Center for Genome Sciences & Systems Biology, E. Martin, B. Koebbe and J. Hoisington-López for technical support and sequencing expertise.
Author information
Authors and Affiliations
Contributions
A.J.G. and G.D. conceived and designed the study. P.I.T., B.B.W. and I.M.N. assembled the cohorts, collected the specimens and biological data and maintained the database, oversaw the transfer of specimens and clinical metadata and provided clinical insights. A.J.G. and B.W. extracted metagenomic DNA from stools and prepared shotgun metagenomic sequencing libraries. A.J.G., B.W. and A.H.-L. performed stool culturing experiments and isolate genomic DNA extraction. A.J.G. and B.W. prepared isolate genome sequencing libraries. E.A.K. performed Enterococcus phenotyping experiments. X.S. created functional metagenomic libraries, performed functional selections and prepared functional metagenomic sequencing libraries. A.J.G. analysed clinical metadata, shotgun metagenomic sequencing data, isolate genome sequencing data and functional metagenomic data. A.J.G. wrote the manuscript with input from G.D., B.B.W. and P.I.T.
Corresponding author
Ethics declarations
Competing interests
P.I.T. is a member of the Scientific Advisory Board of, holds equity in and is a consultant to MediBeacon. P.I.T. is a coinventor on a filed patent application (US Patent application no. 16/200353) to test intestinal permeability in humans that might generate royalty payments. This involvement is not directly relevant to this manuscript. P.I.T. is also a consultant to Takeda Pharmaceuticals on pediatric gastrointestinal disorders and to the Bill & Melinda Gates Foundation on neonatal infections.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7, and Supplementary Tables 2 and 3.
Supplementary Table 1
Clinical and experimental metadata for infants and samples included in this study.
Supplementary Table 4
Statistics for infant gut isolates sequenced in this study.
Rights and permissions
About this article
Cite this article
Gasparrini, A.J., Wang, B., Sun, X. et al. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat Microbiol 4, 2285–2297 (2019). https://doi.org/10.1038/s41564-019-0550-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0550-2
This article is cited by
-
Early life exposure of infants to benzylpenicillin and gentamicin is associated with a persistent amplification of the gut resistome
Microbiome (2024)
-
Patterns of antibiotic administration in Chinese neonates: results from a multi-center, point prevalence survey
BMC Infectious Diseases (2024)
-
Longitudinal dynamics of farmer and livestock nasal and faecal microbiomes and resistomes
Nature Microbiology (2024)
-
Sequence-structure-function characterization of the emerging tetracycline destructase family of antibiotic resistance enzymes
Communications Biology (2024)
-
Baseline azithromycin resistance in the gut microbiota of preterm born infants
Pediatric Research (2024)