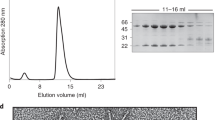
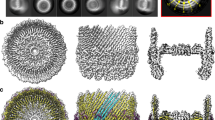
Abstract
Contractile injection systems are sophisticated multiprotein nanomachines that puncture target cell membranes. Although the number of atomic-resolution insights into contractile bacteriophage tails, bacterial type six secretion systems and R-pyocins is rapidly increasing, structural information on the contraction of bacterial phage-like protein-translocation structures directed towards eukaryotic hosts is scarce. Here, we characterize the antifeeding prophage AFP from Serratia entomophila by cryo-electron microscopy. We present the high-resolution structure of the entire AFP particle in the extended state, trace 11 protein chains de novo from the apical cap to the needle tip, describe localization variants and perform specific structural comparisons with related systems. We analyse inter-subunit interactions and highlight their universal conservation within contractile injection systems while revealing the specificities of AFP. Furthermore, we provide the structure of the AFP sheath–baseplate complex in a contracted state. This study reveals atomic details of interaction networks that accompany and define the contraction mechanism of toxin-delivery tailocins, offering a comprehensive framework for understanding their mode of action and for their possible adaptation as biocontrol agents.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The cryo-EM maps and corresponding atomic coordinates have been deposited to the EMDB and PDB with the following accession codes: full extended AFP composite map, EMD-4783; extended AFP baseplate C6 map, EMD-4782 and 6RAO; extended AFP baseplate C3 map, EMD-4800 and 6RBK; extended AFP cap ending in Afp2–Afp16 EMD-4784 and 6RAP; extended AFP cap ending in Afp3–Afp16, EMD-4801; extended AFP sheath map, EMD-4802 and 6RBN; extended AFP needle map from subtracted images, EMD-4871; contracted AFP sheath map, EMD-4803 and 6RC8; contracted AFP baseplate; EMD-4876 and 6RGL and contracted AFP sheath C6 map, EMD-4859. All other data supporting the findings of this study are available from A.D. and I.G. on request.
References
Kube, S. & Wendler, P. Structural comparison of contractile nanomachines. AIMS Biophys. 2, 88–115 (2015).
Taylor, N. M. I., van Raaij, M. J. & Leiman, P. G. Contractile injection systems of bacteriophages and related systems. Mol. Microbiol. 108, 6–15 (2018).
Patz, S. et al. Phage tail-like particles are versatile bacterial nanomachines—a mini-review. J. Adv. Res. 19, 75–84 (2019).
Leiman, P. G. et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl Acad. Sci. USA 106, 4154–4159 (2009).
Brackmann, M., Nazarov, S., Wang, J. & Basler, M. Using force to punch holes: mechanics of contractile nanomachines. Trends Cell Biol. 27, 623–632 (2017).
Nguyen, V. S. et al. Towards a complete structural deciphering of type VI secretion system. Curr. Opin. Struct. Biol. 49, 77–84 (2018).
Nakayama, K. et al. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38, 213–231 (2000).
Michel-Briand, Y. & Baysse, C. The pyocins of Pseudomonas aeruginosa. Biochimie 84, 499–510 (2002).
Ge, P. et al. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat. Struct. Mol. Biol. 22, 377–382 (2015).
Hurst, M. R. H., Glare, T. R. & Jackson, T. A. Cloning Serratia entomophila antifeeding genes—a putative defective prophage active against the grass grub Costelytra zealandica. J. Bacteriol. 186, 5116–5128 (2004).
Yang, G., Dowling, A. J., Gerike, U., Ffrench-Constant, R. H. & Waterfield, N. R. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J. Bacteriol. 188, 2254–2261 (2006).
Sarris, P. F., Ladoukakis, E. D., Panopoulos, N. J. & Scoulica, E. V. A phage tail-derived element withwide distribution among both prokaryotic domains: a comparative genomic and phylogenetic study. Genome Biol. Evol. 6, 1739–1747 (2014).
Heymann, J. B. et al. Three-dimensional structure of the toxin-delivery particle antifeeding prophage of Serratia entomophila. J. Biol. Chem. 288, 25276–25284 (2013).
Jiang, F. et al. Cryo-EM structure and assembly of an extracellular contractile injection system. Cell 177, 370–383 (2019).
Shikuma, N. J. et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343, 529–534 (2014).
Böck, D. et al. In situ architecture, function, and evolution of a contractile injection system. Science 357, 713–717 (2017).
Kudryashev, M. et al. Structure of the type VI secretion system contractile sheath. Cell 160, 952–962 (2015).
Wang, J. et al. Cryo-EM structure of the extended type VI secretion system sheath-tube complex. Nat. Microbiol. 2, 1507–1512 (2017).
Nazarov, S. et al. Cryo‐EM reconstruction of type VI secretion system baseplate and sheath distal end. EMBO J. 37, e97103 (2017).
Taylor, N. M. I. et al. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533, 346–352 (2016).
Hurst, M. R., Glare, T. R., Jackson, T. A. & Ronson, C. W. Plasmid-located pathogenicity determinants of Serratia entomophila, the causal agent of amber disease of grass grub, show similarity to the insecticidal toxins of Photorhabdus luminescens. J. Bacteriol. 182, 5127–5138 (2000).
Rybakova, D. et al. Role of antifeeding prophage (Afp) protein Afp16 in terminating the length of the Afp tailocin and stabilizing its sheath. Mol. Microbiol. 89, 702–714 (2013).
Rybakova, D., Schramm, P., Mitra, A. K. & Hurst, M. R. H. Afp14 is involved in regulating the length of Anti-feeding prophage (Afp). Mol. Microbiol. 96, 815–826 (2015).
Hurst, M. R. H. et al. Serratia proteamaculans strain AGR96X encodes an antifeeding prophage (tailocin) with activity against grass grub (Costelytra giveni) and Manuka beetle (Pyronota species) larvae. Appl. Environ. Microbiol. 84, e02739-17 (2018).
Pell, L. G., Kanelis, V., Donaldson, L. W., Howell, P. L. & Davidson, A. R. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl Acad. Sci. USA 106, 4160–4165 (2009).
Arnaud, C.-A. et al. Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection. Nat. Commun. 8, 1953 (2017).
Biggs, D. R. & McGregor, P. G. Gut pH and amylase and protease activity in larvae of the New Zealand grass grub (Costelytra zealandica; Coleoptera: Scarabiaedae) as a basis for selection inhibitors. Insect Biochem. Mol. Biol. 26, 69–75 (1996).
Bateman, A. & Bycroft, M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299, 1113–1119 (2000).
Zheng, W. et al. Refined cryo-EM structure of the T4 tail tube: exploring the lowest dose limit. Structure 25, 1436–1441 (2017).
Bönemann, G., Pietrosiuk, A., Diemand, A., Zentgraf, H. & Mogk, A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325 (2009).
Zoued, A. et al. Priming and polymerization of a bacterial contractile tail structure. Nature 531, 59–63 (2016).
Zoued, A. et al. TssA: the cap protein of the type VI secretion system tail. Bioessays 39, 1600262 (2017).
Fokine, A. et al. The molecular architecture of the bacteriophage T4 neck. J. Mol. Biol. 425, 1731–1744 (2013).
Pell, L. G. et al. The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J. Mol. Biol. 389, 938–951 (2009).
Cherrak, Y. et al. Biogenesis and structure of a type VI secretion baseplate. Nat. Microbiol. 3, 1404–1416 (2018).
Park, Y.-J. et al. Structure of the type VI secretion system TssK–TssF–TssG baseplate subcomplex revealed by cryo-electron microscopy. Nat. Commun. 9, 5385 (2018).
Shneider, M. M. et al. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353 (2013).
Kanamaru, S., Ishiwata, Y., Suzuki, T., Rossmann, M. G. & Arisaka, F. Control of bacteriophage T4 tail lysozyme activity during the infection process. J. Mol. Biol. 346, 1013–1020 (2005).
Quentin, D. et al. Mechanism of loading and translocation of type VI secretion system effector Tse6. Nat. Microbiol. 3, 1142–1152 (2018).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Desfosses, A., Ciuffa, R., Gutsche, I. & Sachse, C. SPRING - an image processing package for single-particle based helical reconstruction from electron cryomicrographs. J. Struct. Biol. 185, 15–26 (2014).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Egelman, E. H. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Shaikh, T. R. et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974 (2008).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Ilca, S. L. et al. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6, 8843 (2015).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Hoffmann, N. A., Jakobi, A. J., Vorländer, M. K., Sachse, C. & Müller, C. W. Transcribing RNA polymerase III observed by electron cryomicroscopy. FEBS J. 283, 2811–2819 (2016).
Jakobi, A. J., Wilmanns, M. & Sachse, C. Model-based local density sharpening of cryo-EM maps. eLife 6, e27131 (2017).
Kidmose, R. T., Nissen, P., Boesen, T., Karlsen, J. L. & Pedersen, B. P. Namdinator—automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531 (2019).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
This work was supported by a RSNZ Marsden grant to A.K.M. and M.R.H.H. and by the Bio-protection Research Centre. The I.G. lab was funded by a European Union’s Horizon 2020 research and innovation programme under grant agreement no. 647784. A.D. was further supported by the Fondation Recherche Medicale (grant no. ARF20160936266) and Labex GRAL (grant no. ANR-10-LABX-49-01). J.F. was supported by a long-term EMBO fellowship (grant no. ALTF441-2017). M.J. was supported by a doctoral grant from the Commissariat à l’Énergie Atomique et aux Énergies Alternatives (CEA). The authors wish to acknowledge the New Zealand eScience Infrastructure (NeSI) high performance computing facilities—particularly B. Roberts and P. Maxwell who set up the cryo-EM software on the cluster and provided precious support—for the image processing. The national facilities of New Zealand are provided by the NeSI and are jointly funded by collaborator institutions of the NeSI and through the Research Infrastructure programme of the Ministry of Business, Innovation and Employment (https://www.nesi.org.nz). We also acknowledge A. Peuch and G. Schoehn for support and access to the joint IBS/EMBL EM computing cluster, which was used as part of the platforms of the Grenoble Instruct-ERIC center (ISBG; UMS 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology. Platform access was supported by FRISBI (ANR-10-INBS-05-02) and GRAL, a project of the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003). IBS acknowledges integration into the Interdisciplinary Research Institute of Grenoble (IRIG, CEA). We thank A. Jakobi for advice and help with the atomic model refinements and local amplitude scaling of the maps, and for providing early versions of LocScale. We thank A. Turner and the Imaging Centre of the University of Auckland for support and help with the use of the electron microscopes. We thank C. Sachse for help and advice in image processing and in setting SPRING on the NeSI and the IBS/EMBL clusters. We thank M. Middleditch for the mass-spectrometry experiments and help with the data analysis. The EMBL Cryo-Electron Microscopy Service Platform is acknowledged for support in image acquisition and analysis. We are grateful to W. Hagen for the acquisition of high-quality cryo-EM images of extended AFP in 2016. Data acquisition has been supported by iNEXT (grant no. 653706) funded by the EU Horizon 2020 programme. We acknowledge P. Leiman for critical comments of the manuscript.
Author information
Authors and Affiliations
Contributions
A.K.M. and M.R.H.H. designed and funded the project. A.D., H.V. and M.J. purified and initially characterized samples by negative staining and cryo-EM. A.D. analysed the mass-spectrometry data. A.D. performed the image analyses with significant input from H.V. A.D. performed the atomic model building with significant input from H.V. and J.F. J.H. and H.J. collected the high-resolution cryo-EM datasets of the contracted AFP. A.D., H.V., T.J., J.F., M.J., J.B.H., M.R.H.H., I.G. and A.K.M. analysed the data. A.D., J.F., M.J. and I.G. prepared the figures and tables. A.K.M., M.R.H.H. and I.G. provided support and supervised the project at various stages. I.G. and A.K.M. wrote the manuscript with significant input from M.R.H.H., J.B.H., A.D., J.F. and M.J., and contributions from all of the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9 and Supplementary Tables 1–6.
Rights and permissions
About this article
Cite this article
Desfosses, A., Venugopal, H., Joshi, T. et al. Atomic structures of an entire contractile injection system in both the extended and contracted states. Nat Microbiol 4, 1885–1894 (2019). https://doi.org/10.1038/s41564-019-0530-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0530-6
This article is cited by
-
An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano
Nature Communications (2024)
-
Programmable protein delivery with a bacterial contractile injection system
Nature (2023)
-
Cytoplasmic contractile injection systems mediate cell death in Streptomyces
Nature Microbiology (2023)
-
High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217
Nature Communications (2023)
-
A contractile injection system is required for developmentally regulated cell death in Streptomyces coelicolor
Nature Communications (2023)