Abstract
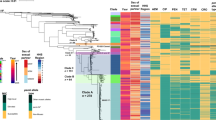
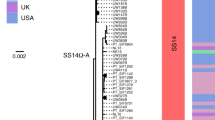
The sexually transmitted pathogen Neisseria gonorrhoeae is regarded as being on the way to becoming an untreatable superbug. Despite its clinical importance, little is known about its emergence and evolution, and how this corresponds with the introduction of antimicrobials. We present a genome-based phylogeographical analysis of 419 gonococcal isolates from across the globe. Results indicate that modern gonococci originated in Europe or Africa, possibly as late as the sixteenth century and subsequently disseminated globally. We provide evidence that the modern gonococcal population has been shaped by antimicrobial treatment of sexually transmitted infections as well as other infections, leading to the emergence of two major lineages with different evolutionary strategies. The well-described multidrug-resistant lineage is associated with high rates of homologous recombination and infection in high-risk sexual networks. A second, multisusceptible lineage is more associated with heterosexual networks, with potential implications for infection control.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All genomic data have been deposited in the European Nucleotide Archive (ENA) under project number PRJEB4024. Accession numbers for the particular strains are indicated in Supplementary Table 1. All other data supporting the findings of this study are available within the paper and its Supplementary Information files.
Code availability
The custom Perl script to convert xmfa to fasta files (xmfa2fas.pl) is available from https://gist.github.com/leosanbu/.
References
Newman, L. et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS ONE 10, e0143304 (2015).
Unemo, M., Del Rio, C. & Shafer, W. M. Antimicrobial resistance expressed by Neisseria gonorrhoeae: a major global public health problem in the 21st century. Microbiol. Spectr. 4, EI10-0009-2015 (2016).
Fifer, H. et al. Failure of dual antimicrobial therapy in treatment of gonorrhea. N. Engl. J. Med. 374, 2504–2506 (2016).
Wi, T. et al. Antimicrobial resistance in Neisseria gonorrhoeae: global surveillance and a call for international collaborative action. PLoS Med. 14, e1002344 (2017).
Fingerhuth, S. M., Bonhoeffer, S., Low, N. & Althaus, C. L. Antibiotic-resistant Neisseria gonorrhoeae spread faster with more treatment, not more sexual partners. PLoS Pathog. 12, e1005611 (2016).
Annual Epidemiological Report for 2016 (ECDC, 2018); https://ecdc.europa.eu/en/publications-data/gonorrhoea-annual-epidemiological-report-2016
Luys, G. A Textbook on Gonorrhoea and its Complications (Baillière, Tindall and Cox, 1913).
Grmek, M. Diseases in the Ancient Greek World (John Hopkins Univ. Press, 1989).
Oriel, J. D. The Scars of Venus: A History of Venereology (Springer, 1994).
Spratt, B. G. Hybrid penicillin-binding proteins in penicillin-resistant strains of Neisseria gonorrhoeae. Nature 332, 173–176 (1988).
Ohnishi, M. et al. Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea?: Detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrob. Agents Chemother. 55, 3538–3545 (2011).
Ohnishi, M. Ceftriaxone-resistant Neisseria gonorrhoeae, Japan. Emerg. Infect. Dis. 17, 148–149 (2011).
Lefebvre, B. et al. Ceftriaxone-resistant Neisseria gonorrhoeae, Canada, 2017. Emerg. Infect. Dis. 24, 381–383 (2018).
Golparian, D. et al. Multidrug-resistant Neisseria gonorrhoeae isolate, belonging to the internationally spreading Japanese FC428 clone, with ceftriaxone resistance and intermediate resistance to azithromycin, Ireland, August 2018. Euro Surveill. 23, 1800617 (2018).
Popovich, K. J. & Snitkin, E. S. Whole genome sequencing—implications for infection prevention and outbreak investigations. Curr. Infect. Dis. Rep. 19, 15 (2017).
Grad, Y. H. et al. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect. Dis. 14, 220–226 (2014).
Demczuk, W. et al. Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J. Clin. Microbiol. 53, 191–200 (2015).
De Silva, D. et al. Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect. Dis. 16, 1295–1303 (2016).
Didelot, X. et al. Genomic analysis and comparison of two gonorrhea outbreaks. mBio 7, e00525-16 (2016).
Chisholm, S. A. et al. An outbreak of high-level azithromycin resistant Neisseria gonorrhoeae in England. Sex Transm. Infect. 92, 365–367 (2016).
Duchene, S. et al. Genome-scale rates of evolutionary change in bacteria. Microb. Genom. 2, e000094 (2016).
Demczuk, W. et al. Neisseria gonorrhoeae sequence typing for antimicrobial resistance, a novel antimicrobial resistance multilocus typing scheme for tracking global dissemination of N. gonorrhoeae strains. J. Clin. Microbiol. 55, 1454–1468 (2017).
Cámara, J. et al. Molecular characterization of two high-level ceftriaxone-resistant Neisseria gonorrhoeae isolates detected in Catalonia, Spain. J. Antimicrob. Chemother. 67, 1858–1860 (2012).
Unemo, M. et al. High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob. Agents Chemother. 56, 1273–1280 (2012).
Flett, F., Humphreys, G. O. & Saunders, J. R. Intraspecific and intergeneric mobilization of non-conjugative resistance plasmids by a 24.5 megadalton conjugative plasmid of Neisseria gonorrhoeae. J. Gen. Microbiol. 125, 123–129 (1981).
Demczuk, W. et al. Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J. Clin. Microbiol. 54, 1304–1313 (2016).
Harris, S. R. et al. Public health surveillance of multidrug-resistant clones of Neisseria gonorrhoeae in Europe: a genomic survey. Lancet Infect. Dis. 18, 758–768 (2018).
Hopkins, A. G. Globalization in World History (Pimlico, 2002).
McNeill, W. H. Plagues and Peoples (Doubleday, 1977).
Huovinen, P. Resistance to trimethoprim–sulfamethoxazole. Clin. Infect. Dis. 32, 1608–1614 (2001).
Workowski, K. A. & Bolan, G. A. Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm. Rep. 64, 1–137 (2015).
Kenyon, C. R. & Schwartz, I. S. Effects of sexual network connectivity and antimicrobial drug use on antimicrobial resistance in Neisseria gonorrhoeae. Emerg. Infect. Dis. 24, 1195–1203 (2018).
Turner, K. M. & Garnett, G. P. The impact of the phase of an epidemic of sexually transmitted infection on the evolution of the organism. Sex. Transm. Infect. 78, i20–i30 (2002).
Wilkinson, D. et al. Unrecognized sexually transmitted infections in rural South African women: a hidden epidemic. Bull. World Health Organ. 77, 22–28 (1999).
Hazel, A., Ponnaluri-Wears, S., Davis, G. S., Low, B. S. & Foxman, B. High prevalence of Neisseria gonorrhoeae in a remote, undertreated population of Namibian pastoralists. Epidemiol. Infect. 142, 2422–2432 (2014).
Grad, Y. H. et al. Genomic epidemiology of gonococcal resistance to extended spectrum cephalosporins, macrolides, and fluoroquinolones in the US, 2000–2013. J. Infect. Dis. 214, 1579–1587 (2016).
Unemo, M., Olcen, P., Berglund, T., Albert, J. & Fredlund, H. Molecular epidemiology of Neisseria gonorrhoeae: sequence analysis of the porB gene confirms presence of two circulating strains. J. Clin. Microbiol. 40, 3741–3749 (2002).
Unemo, M., Fasth, O., Fredlund, H., Limnios, A. & Tapsall, J. Phenotypic and genetic characterization of the 2008 WHO Neisseria gonorrhoeae reference strain panel intended for global quality assurance and quality control of gonococcal antimicrobial resistance surveillance for public health purposes. J. Antimicrob. Chemother. 63, 1142–1151 (2009).
Li, H. et al. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Harris, S. R. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474 (2010).
Darling, A. E., Mau, B. & Perna, N. T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5, e11147 (2010).
Piekarowicz, A. et al. Characterization of the dsDNA prophage sequences in the genome of Neisseria gonorrhoeae and visualization of productive bacteriophage. BMC Microbiol. 7, 66 (2007).
Croucher, N. J. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15 (2015).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Page, A. J. et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2, e000056 (2016).
Cheng, L., Connor, T. R., Siren, J., Aanensen, D. M. & Corander, J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 30, 1224–1228 (2013).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Lemoine, F. et al. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556, 452–456 (2018).
Page, A. J. et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb. Genom. 2, e000083 (2016).
Jolley, K. A. & Maiden, M. C. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (2010).
Martin, I. M., Ison, C. A., Aanensen, D. M., Fenton, K. A. & Spratt, B. G. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J. Infect. Dis. 189, 1497–1505 (2004).
Kwong, J. C. et al. NGMASTER: in silico multi-antigen sequence typing for Neisseria gonorrhoeae. Microb. Genom. 2, e000076 (2016).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Hunt, M. et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genom. 3, e000131 (2017).
Turner, A., Gough, K. R. & Leeming, J. P. Molecular epidemiology of tetM genes in Neisseria gonorrhoeae. Sex. Transm. Infect. 75, 60–66 (1999).
Dillon, J. R., Li, H., Yeung, K. & Aman, T. A. A PCR assay for discriminating Neisseria gonorrhoeae β-lactamase-producing plasmids. Mol. Cell. Probes 13, 89–92 (1999).
Kamvar, Z. N., Tabima, J. F. & Grunwald, N. J. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281 (2014).
Excoffier, L., Smouse, P. E. & Quattro, J. M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491 (1992).
Dray, S. & Dufour, A. B. The ade4 package: implementing the duality diagram for ecologists. J. Stat. Softw. 22, 1–20 (2007).
Jombart, T., Devillard, S. & Balloux, F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11, 94 (2010).
Jombart, T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
Louis, T. A. Multivariate analysis of variance and repeated measures, a practical approach for behavioural scientists. D. J. Hand and C. C. Taylor, Chapman & Hall, London. Stat. Med. https://doi.org/10.1002/sim.4780081218 (1989).
Benjamini, Y. Discovering the false discovery rate. J. R. Stat. Soc. Series B Stat. Methodol. 72, 405–416 (2010).
Murray, G. G. et al. The effect of genetic structure on molecular dating and tests for temporal signal. Methods Ecol. Evol. 7, 80–89 (2016).
Duchene, S., Duchene, D., Holmes, E. C. & Ho, S. Y. The performance of the date-randomization test in phylogenetic analyses of time-structured virus data. Mol. Biol. Evol. 32, 1895–1906 (2015).
To, T. H., Jung, M., Lycett, S. & Gascuel, O. Fast dating using least-squares criteria and algorithms. Syst. Biol. 65, 82–97 (2016).
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Ansari, M. A. & Didelot, X. Bayesian inference of the evolution of a phenotype distribution on a phylogenetic tree. Genetics 204, 89–98 (2016).
Huelsenbeck, J. P., Nielsen, R. & Bollback, J. P. Stochastic mapping of morphological characters. Syst. Biol. 52, 131–158 (2003).
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012).
Bollback, J. P. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7, 88 (2006).
Unemo, M. & Shafer, W. M. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin. Microbiol. Rev. 27, 587–613 (2014).
R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2008).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
Hadfield, J. et al. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34, 292–293 (2018).
Acknowledgements
We thank H. To and O. Gascuel for their help with the LSD software, and the Pathogen Informatics group at the Wellcome Sanger Institute for informatics support. We also thank S. Szreter, T. Bayliss-Smith and P. Mitchell from the University of Cambridge, Cambridge, UK, for interesting discussions on the historical evidence of gonorrhoea infection. The Japanese isolates were kindly provided by Y. Watanabe and T. Kuroki, Department of Microbiology, Kanagawa Prefectural Institute of Public Health, Kanagawa, Japan. This work was funded by the Wellcome grant number 098051 and the Foundation for Medical Research at Örebro University Hospital, Örebro, Sweden. J.C. was funded by the ERC grant number 745258. Y.H.G. is supported by The Smith Family Foundation and the NIH/NIAID grant 1R01AI132606-01.
Author information
Authors and Affiliations
Contributions
S.R.H., M.U., S.D.B. and J.P. conceived and managed the study. L.S.B. and S.R.H. analysed the data and drafted the manuscript. D.G., M.U. and M.O. cultured isolates and extracted DNA. L.S.B., S.R.H., M.U. and Y.H.G. interpreted the data. J.C. provided statistical analysis. R.F. advised on historical interpretation. All authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10, Supplementary Tables 2 and 3, Supplementary Tables 5–7 and Supplementary Notes.
Supplementary Table 1
Additional strain information.
Supplementary Table 4
Prior and posterior continent assignments.
Rights and permissions
About this article
Cite this article
Sánchez-Busó, L., Golparian, D., Corander, J. et al. The impact of antimicrobials on gonococcal evolution. Nat Microbiol 4, 1941–1950 (2019). https://doi.org/10.1038/s41564-019-0501-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0501-y
This article is cited by
-
CanB is a metabolic mediator of antibiotic resistance in Neisseria gonorrhoeae
Nature Microbiology (2023)
-
Emergence and evolution of antimicrobial resistance genes and mutations in Neisseria gonorrhoeae
Genome Medicine (2021)
-
Antimicrobial resistance and molecular epidemiological typing of Neisseria gonorrhoeae isolates from Kyrgyzstan in Central Asia, 2012 and 2017
BMC Infectious Diseases (2021)
-
Epidemiology, Treatments, and Vaccine Development for Antimicrobial-Resistant Neisseria gonorrhoeae: Current Strategies and Future Directions
Drugs (2021)
-
A community-driven resource for genomic epidemiology and antimicrobial resistance prediction of Neisseria gonorrhoeae at Pathogenwatch
Genome Medicine (2021)