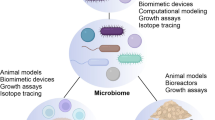
Abstract
Advances in metagenome sequencing of the human microbiome have provided a plethora of new insights and revealed a close association of this complex ecosystem with a range of human diseases. However, there is little knowledge about how the different members of the microbial community interact with each other and with the host, and we lack basic mechanistic understanding of these interactions related to health and disease. Mathematical modelling has been demonstrated to be highly advantageous for gaining insights into the dynamics and interactions of complex systems and in recent years, several modelling approaches have been proposed to enhance our understanding of the microbiome. Here, we review the latest developments and current approaches, and highlight how different modelling strategies have been applied to unravel the highly dynamic nature of the human microbiome. Furthermore, we discuss present limitations of different modelling strategies and provide a perspective of how modelling can advance understanding and offer new treatment routes to impact human health.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lozupone, C., Stombaugh, J., Gordon, J. & Jansson, J. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Gilbert, J. A. et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 535, 94–103 (2016).
Van Wey, A. S. et al. Monoculture parameters successfully predict coculture growth kinetics of Bacteroides thetaiotaomicron and two Bifidobacterium strains. Int. J. Food Microbiol. 191, 172–181 (2014).
Muñoz-Tamayo, R. et al. Kinetic modelling of lactate utilization and butyrate production by key human colonic bacterial species. FEMS Microbiol. Ecol. 76, 615–624 (2011).
White, R. A., Callister, S. J., Moore, R. J., Baker, E. S. & Jansson, J. K. The past, present and future of microbiome analyses. Nat. Protoc. 11, 2049–2053 (2016).
Arnold, J. W., Roach, J. & Azcarate-Peril, M. A. Emerging technologies for gut microbiome research. Trends Microbiol. 24, 887–901 (2016).
Amaretti, A. et al. Kinetics and metabolism of Bifidobacterium adolescentis MB 239 growing on glucose, galactose, lactose, and galactooligosaccharides. Appl. Environ. Microbiol. 73, 3637–3644 (2007).
Lagier, J. C. et al. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 28, 237–264 (2015).
Tomlin, C. J. & Axelrod, J. D. Biology by numbers: mathematical modelling in developmental biology. Nat. Rev. Genet. 8, 331–340 (2007).
Fabien, B. Analytical System Dynamics (Springer, 2009).
Van Wey, A. S., Lovatt, S. J., Roy, N. C. & Shorten, P. R. Determination of potential metabolic pathways of human intestinal bacteria by modeling growth kinetics from cross-feeding dynamics. Food Res. Int. 88, 207–216 (2016).
Muñoz-Tamayo, R., Laroche, B., Walter, É., Doré, J. & Leclerc, M. Mathematical modelling of carbohydrate degradation by human colonic microbiota. J. Theor. Biol. 266, 189–201 (2010).
Vandeputte, D. et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65, 57–62 (2016).
Vieira-Silva, S. & Rocha, E. P. C. The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet. 6, e1000808 (2010).
Stein, R. R. et al. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput. Biol. 9, 31–36 (2013).
Nielsen, H. B. et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 32, 822–828 (2014).
Mounier, J. et al. Microbial interactions within a cheese microbial community. Appl. Environ. Microbiol. 74, 172–181 (2008).
Korem, T. et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101–1106 (2015).
Faust, K. et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 8, e1002606 (2012).
Shoaie, S. et al. Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci. Rep. 3, 2532 (2013).
El-Semman, I. E. et al. Genome-scale metabolic reconstructions of Bifidobacterium adolescentis L2–32 and Faecalibacterium prausnitzii A2–165 and their interaction. BMC Syst. Biol. 8, 41 (2014).
Steinway, S. N., Biggs, M. B., Loughran, T. P., Papin, J. A. & Albert, R. Inference of network dynamics and metabolic interactions in the gut microbiome. PLoS Comput. Biol. 11, e1004338 (2015).
Shoaie, S. et al. Quantifying diet-induced metabolic changes of the human gut microbiome. Cell Metab. 22, 320–331 (2015).
Heinken, A. & Thiele, I. Anoxic conditions promote species-specific mutualism between gut microbes in silico. Appl. Environ. Microbiol. 81, 4049–4061 (2015).
Lidicker, W. Z. A clarification of interactions in ecological systems. Bioscience 29, 475–477 (1979).
Magnúsdóttir, S. et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 35, 81–89 (2016).
Zelezniak, A. et al. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl Acad. Sci. USA 112, 6449–6454 (2015).
Karlsson, F. H., Nookaew, I., Petranovic, D. & Nielsen, J. Prospects for systems biology and modeling of the gut microbiome. Trends Biotechnol. 29, 251–258 (2011).
Bonabeau, E. Agent-based modeling: methods and techniques for simulating human systems. Proc. Natl Acad. Sci. USA 99, 7280–7287 (2002).
Shashkova, T. et al. Agent based modeling of human gut microbiome interactions and perturbations. PLoS ONE 11, e0148386 (2016).
Pinto, F., Medina, D. A., Pérez-Correa, J. R. & Garrido, D. Modeling metabolic interactions in a consortium of the infant gut microbiome. Front. Microbiol. 8, 2507 (2017).
Ruan, Q. et al. Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 22, 2532–2538 (2006).
Deng, Y. et al. Molecular ecological network analyses. BMC Bioinformatics 13, 133 (2012).
Hoffmann, C. et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE 8, e66019 (2013).
Dice, L. R. Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–109 (2011).
Friedman, J. & Alm, E. J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8, e1002687 (2012).
Kurtz, Z. D. et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 11, e1004226 (2015).
Faust, K. & Raes, J. CoNet app: inference of biological association networks using Cytoscape. F1000 5, 1519 (2016).
Faust, K. & Raes, J. Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550 (2012).
Meinshausen, N. & Bühlmann, P. High-dimensional graphs and variable selection with the Lasso. Ann. Stat. 34, 1436–1462 (2006).
Bonneau, R. et al. The inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 7, R36 (2006).
Friedman, J., Hastie, T. & Tibshirani, R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics 9, 432–441 (2008).
Banerjee, O. & Ghaoui, L. El & D’Aspremont, A. Model selection through sparse maximum likelihood estimation for multivariate gaussian or binary data. J. Mach. Learn. Res. 9, 485–516 (2008).
Sarkar, S. K. & Chang, C. K. The simes method for multiple hypothesis testing with positively dependent test statistics. J. Am. Stat. Assoc. 92, 1601–1608 (1997).
Vandeputte, D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017).
Marino, S., Baxter, N. T., Huffnagle, G. B., Petrosino, J. F. & Schloss, P. D. Mathematical modeling of primary succession of murine intestinal microbiota. Proc. Natl Acad. Sci. USA 111, 439–444 (2014).
Weiss, S. et al. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 10, 1669–1681 (2016).
Venturelli, O. S. et al. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol. 14, e8157 (2018).
Faust, K. et al. Signatures of ecological processes in microbial community time series. Microbiome 6, 120 (2018).
Bucci, V. & Xavier, J. B. Towards predictive models of the human gut microbiome. J. Mol. Biol. 426, 3907–3916 (2014).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Brown, C. T., Olm, M. R., Thomas, B. C. & Banfield, J. F. Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 34, 1256–1263 (2016).
Greenblum, S., Turnbaugh, P. J. & Borenstein, E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc. Natl Acad. Sci. USA 109, 594–599 (2011).
Chan, S. H. J., Simons, M. N. & Maranas, C. D. SteadyCom: predicting microbial abundances while ensuring community stability. PLoS Comput. Biol. 13, e1005539 (2017).
Shoaie, S. et al. Quantifying diet-induced metabolic changes of the human gut microbiome. Cell Metab. 22, 320–331 (2015).
Zengler, K. & Palsson, B. O. A road map for the development of community systems (CoSy) biology. Nat. Rev. Microbiol. 10, 366–372 (2012).
Oberhardt, M. A., Palsson, B. & Papin, J. A. Applications of genome-scale metabolic reconstructions. Mol. Syst. Biol. 5, 320 (2009).
Lewis, N. E., Nagarajan, H. & Palsson, B. O. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 10, 291–305 (2012).
Orth, J. D., Thiele, I. & Palsson, B. Ø. What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010).
Levy, S. E. & Myers, R. M. Advancements in next-generation sequencing. Annu. Rev. Genom. Hum. Genet. 17, 95–115 (2016).
Heinken, A., Sahoo, S., Fleming, R. M. T. & Thiele, I. Systems-level characterization of a host–microbe metabolic symbiosis in the mammalian gut. Gut Microbes 4, 28–40 (2013).
Henry, C. S. et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977–982 (2010).
Stolyar, S. et al. Metabolic modeling of a mutualistic microbial community. Mol. Syst. Biol. 3, 92 (2007).
Zomorrodi, A. R. & Maranas, C. D. OptCom: A multi-level optimization framework for the metabolic modeling and analysis of microbial communities. PLoS Comput. Biol. 8, e1002363 (2012).
Khandelwal, R. A., Olivier, B. G., Röling, W. F. M., Teusink, B. & Bruggeman, F. J. Community flux balance analysis for microbial consortia at balanced growth. PLoS ONE 8, e64567 (2013).
Lozupone, C., Stomabaugh, J., Gordon, J., Jansson, J. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Caporaso, J. G. et al. Moving pictures of the human microbiome. Genome Biol. 12, R50 (2011).
David, L. A. et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2015).
Faith, J. J. et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013).
Kumar, M. et al. Gut microbiota dysbiosis is associated with malnutrition and reduced plasma amino acid levels: Lessons from genome-scale metabolic modeling. Metab. Eng. 49, 128–142 (2018).
Babaei, P., Shoaie, S., Ji, B. & Nielsen, J. Challenges in modeling the human gut microbiome. Nat. Biotechnol. 36, 682–686 (2018).
Garza, D. R., Van Verk, M. C., Huynen, M. A. & Dutilh, B. E. Towards predicting the environmental metabolome from metagenomics with a mechanistic model. Nat. Microbiol. 3, 456–460 (2018).
Diener, C. & Resendis-Antonio, O. Micom: metagenome-scale modeling to infer metabolic interactions in the microbiota. Preprint at https://www.biorxiv.org/content/10.1101/361907v2 (2018).
Harrison, R., Papp, B., Pál, C., Oliver, S. G. & Delneri, D. Plasticity of genetic interactions in metabolic networks of yeast. Proc. Natl Acad. Sci. USA 104, 2307–2312 (2007).
Szappanos, B. et al. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat. Genet. 43, 656–662 (2011).
Mani, R., St.Onge, R. P., Hartman, J. L., Giaever, G. & Roth, F. P. Defining genetic interaction. Proc. Natl Acad. Sci. USA 105, 3461–3466 (2008).
Coyte, K. Z., Schluter, J. & Foster, K. R. The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666 (2015).
Bauer, E., Zimmermann, J., Baldini, F., Thiele, I. & Kaleta, C. BacArena: individual-based metabolic modeling of heterogeneous microbes in complex communities. PLoS Comput. Biol. 13, e1005544 (2017).
Glushchenko, O. et al. VERA: agent-based modeling transmission of antibiotic resistance between human pathogens and gut microbiota. Bioinformatics https://doi.org/10.1093/bioinformatics/btz154 (2019).
Weston, B., Fogal, B., Cook, D. & Dhurjati, P. An agent-based modeling framework for evaluating hypotheses on risks for developing autism: effects of the gut microbial environment. Med. Hypotheses 84, 395–401 (2015).
An, G., Mi, Q., Dutta-Moscato, J. & Vodovotz, Y. Agent-based models in translational systems biology. WIREs Syst. Biol. Med. 1, 159–171 (2009).
Mardinoglu, A. et al. The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol. Syst. Biol. 11, 834–834 (2015).
Arora, T. et al. Diabetes-associated microbiota in fa/fa rats is modified by Roux-en-Y gastric bypass. ISME J. 11, 2035–2046 (2017).
Arora, T. et al. Microbially produced glucagon-like peptide 1 improves glucose tolerance in mice. Mol. Metab. 5, 725–730 (2016).
Darzi, Y., Falony, G., Vieira-Silva, S. & Raes, J. Towards biome-specific analysis of meta-omics data. ISME J. 10, 1025–1028 (2016).
Wang, Y., Eddy, J. A. & Price, N. D. Reconstruction of genome-scale metabolic models for 126 human tissues using mCADRE. BMC Syst. Biol. 6, 153 (2012).
Agren, R. et al. Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput. Biol. 8, e1002518 (2012).
Schultz, A. & Qutub, A. A. Reconstruction of tissue-specific metabolic networks using CORDA. PLoS Comput. Biol. 12, e1004808 (2016).
Zhalnina, K., Zengler, K., Newman, D. & Northen, T. R. Need for laboratory ecosystems to unravel the structures and functions of soil microbial communities mediated by chemistry. mBio 9, e01175–18 (2018).
Petriz, B. A. & Franco, O. L. Metaproteomics as a complementary approach to gut microbiota in health and disease. Front. Chem. 5, 4 (2017).
de Haffmann, E. Tandem mass spectrometry: a primer. J. Mass Spectrom. 31, 129–137 (1996).
Benesty, J., Chen, J., Huang, Y. & Cohen, I. Noise reduction in speech processing (Springer, 2009).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Huson, D., Auch, A., Qi, J. & Schuster, S. MEGAN analysis of metagenome data. Genome Res. 17, 377–386 (2007).
Karlsson, F. H., Nookaew, I. & Nielsen, J. Metagenomic data utilization and analysis (MEDUSA) and construction of a global gut microbial gene catalogue. PLoS Comput. Biol. 10, e1003706 (2014).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Meyer, F. et al. The metagenomics RAST server — a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008).
Kultima, J. R. et al. MOCAT: A metagenomics assembly and gene prediction toolkit. PLoS ONE 7, e47656 (2012).
Kultima, J. R. et al. MOCAT2: A metagenomic assembly, annotation and profiling framework. Bioinformatics 32, 2520–2523 (2016).
Oulas, A. et al. Metagenomics: tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinform. Biol. Insights 9, 75–88 (2015).
Kunin, V., Copeland, A., Lapidus, A., Mavromatis, K. & Hugenholtz, P. A bioinformatician’s guide to metagenomics. Microbiol. Mol. Biol. Rev. 72, 557–578 (2008).
Escobar-Zepeda, A. et al. Analysis of sequencing strategies and tools for taxonomic annotation: defining standards for progressive metagenomics. Sci. Rep. 8, 12034 (2018).
Lillacci, G. & Khammash, M. Parameter estimation and model selection in computational biology. PLoS Comput. Biol. 6, e1000696 (2010).
Thiele, I. & Palsson, B. Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5, 93–121 (2010).
Chan, S. H. J., Cai, J., Wang, L., Simons-Senftle, M. N. & Maranas, C. D. Standardizing biomass reactions and ensuring complete mass balance in genome-scale metabolic models. Bioinformatics 33, 3603–3609 (2017).
Henry, C. S., Jankowski, M. D., Broadbelt, L. J. & Hatzimanikatis, V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys. J. 90, 1453–1461 (2006).
Agren, R. et al. The RAVEN Toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput. Biol. 9, e1002980 (2013).
Feist, A. M. & Palsson, B. O. The biomass objective function. Curr. Opin. Microbiol. 13, 344–349 (2010).
Mahadevan, R. & Schilling, C. H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5, 264–276 (2003).
Tian, M. & Reed, J. L. Integrating proteomic or transcriptomic data into metabolic models using linear bound flux balance analysis. Bioinformatics 34, 3882–3888 (2018).
Sung, J., Hale, V., Merkel, A. C., Kim, P. J. & Chia, N. Metabolic modeling with big data and the gut microbiome. Appl. Transl. Genom. 10, 10–15 (2016).
Levy, R. & Borenstein, E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl Acad. Sci. USA 110, 12804–12809 (2013).
Embree, M., Liu, J. K., Al-Bassam, M. M. & Zengler, K. Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc. Natl Acad. Sci. USA 112, 15450–15455 (2015).
Nagarajan, H. et al. Characterization and modelling of interspecies electron transfer mechanisms and microbial community dynamics of a syntrophic association. Nat. Commun. 4, 2809 (2013).
Karp, P. D., Paley, S. & Romero, P. The pathway tools software. Bioinformatics 18, 225–232 (2002).
Arakawa, K., Yamada, Y., Shinoda, K., Nakayama, Y. & Tomita, M. GEM system: automatic prototyping of cell-wide metabolic pathway models from genomes. BMC Bioinformatics 7, 168 (2006).
Pitkänen, E. et al. Comparative genome-scale reconstruction of gapless metabolic networks for present and ancestral species. PLoS Comput. Biol. 10, e1003465 (2014).
Dias, O., Rocha, M., Ferreira, E. C. & Rocha, I. Reconstructing high-quality large-scale metabolic models with merlin. Methods Mol. Biol. 1716, 1–36 (2018).
Machado, D., Andrejev, S., Tramontano, M. & Patil, K. R. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 46, 7542–7553 (2018).
Swainston, N. et al. The SuBliMinaL Toolbox: automating steps in the reconstruction of metabolic networks. J. Integr. Bioinform. 8, 186 (2011).
Becker, S. A. et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat. Protoc. 2, 727–738 (2007).
Ebrahim, A., Palsson, J. A. L. B. O. & Hyduke, D. R. COBRApy: constraints-based reconstruction and analysis for Python. BMC Syst. Biol. 7, 74 (2013).
Heirendt, L., Thiele, I. & Fleming, R. M. T. DistributedFBA.jl: high-level, high-performance flux balance analysis in Julia. Bioinformatics 33, 1421–1423 (2017).
Olivier, B. G., Rohwer, J. M. & Hofmeyr, J. H. S. Modelling cellular systems with PySCeS. Bioinformatics 21, 560–561 (2005).
Gelius-Dietrich, G., Desouki, A. A., Fritzemeier, C. J. & Lercher, M. J. Sybil-efficient constraint-based modelling in R. BMC Syst. Biol. 7, 125 (2013).
Seif, Y. et al. Genome-scale metabolic reconstructions of multiple Salmonella strains reveal serovar-specific metabolic traits. Nat. Commun. 9, 3771 (2018).
Monk, J. M. et al. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc. Natl Acad. Sci. USA 110, 20338–20343 (2013).
Kang, D. D., Froula, J., Egan, R. & Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3, e1165 (2015).
Imelfort, M. et al. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ 2, e603 (2014).
Tramontano, M. et al. Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat. Microbiol. 3, 514–522 (2018).
Lieven, C. et al. Memote: A community driven effort towards a standardized genome-scale metabolic model test suite. Preprint at https://www.biorxiv.org/content/10.1101/350991v1 (2018).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
Schellenberger, J., Park, J. O., Conrad, T. M. & Palsson, B. Ø. BiGG: a biochemical genetic and genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics 11, 213 (2010).
Caspi, R. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 44, D471–D480 (2016).
Benedict, M. N., Mundy, M. B., Henry, C. S., Chia, N. & Price, N. D. Likelihood-based gene annotations for gap filling and quality assessment in genome-scale metabolic models. PLoS Comput. Biol. 10, e1003882 (2014).
Latendresse, M. Efficiently gap-filling reaction networks. BMC Bioinformatics 15, 225 (2014).
Acknowledgements
We acknowledge financial support from Knut and Alice Wallenberg Foundation, the Novo Nordisk Foundation (grant no. NNF10CC1016517), Vetenskapsrådet, Bill & Melinda Gates Foundation (grant no. OPP1127499), MetaCardis (grant no. HEALTH-F4-2012-305312), FORMAS and the Swedish Foundation for Strategic Research.
Author information
Authors and Affiliations
Contributions
M.K., B.J. and J.N. collectively conceptualized the manuscript. M.K., B.J., K.Z. and J.N. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumar, M., Ji, B., Zengler, K. et al. Modelling approaches for studying the microbiome. Nat Microbiol 4, 1253–1267 (2019). https://doi.org/10.1038/s41564-019-0491-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0491-9
This article is cited by
-
Gut microbiome-metabolome interactions predict host condition
Microbiome (2024)
-
Predictions of rhizosphere microbiome dynamics with a genome-informed and trait-based energy budget model
Nature Microbiology (2024)
-
Designing a synthetic microbial community through genome metabolic modeling to enhance plant–microbe interaction
Environmental Microbiome (2023)
-
Metagenome-based metabolic modelling predicts unique microbial interactions in deep-sea hydrothermal plume microbiomes
ISME Communications (2023)
-
A mathematical model of Bacteroides thetaiotaomicron, Methanobrevibacter smithii, and Eubacterium rectale interactions in the human gut
Scientific Reports (2023)