Abstract
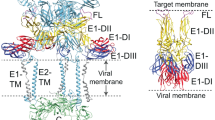
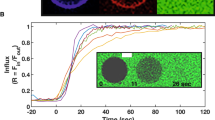
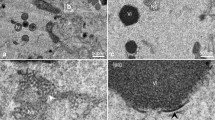
To achieve efficient binding and subsequent fusion, most enveloped viruses encode between one and five proteins1. For many viruses, the clustering of fusion proteins—and their distribution on virus particles—is crucial for fusion activity2,3. Poxviruses, the most complex mammalian viruses, dedicate 15 proteins to binding and membrane fusion4. However, the spatial organization of these proteins and how this influences fusion activity is unknown. Here, we show that the membrane of vaccinia virus is organized into distinct functional domains that are critical for the efficiency of membrane fusion. Using super-resolution microscopy and single-particle analysis, we found that the fusion machinery of vaccinia virus resides exclusively in clusters at virion tips. Repression of individual components of the fusion complex disrupts fusion-machinery polarization, consistent with the reported loss of fusion activity5. Furthermore, we show that displacement of functional fusion complexes from virion tips disrupts the formation of fusion pores and infection kinetics. Our results demonstrate how the protein architecture of poxviruses directly contributes to the efficiency of membrane fusion, and suggest that nanoscale organization may be an intrinsic property of these viruses to assure successful infection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding authors on reasonable request.
Code availability
All custom code used for analysis in the current study is available on GitHub at https://github.com/HenriquesLab/fusiontools.
References
Kielian, M. & Rey, F. A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4, 67–76 (2006).
Harrison, S. C. Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008).
White, J. M., Delos, S. E., Brecher, M. & Schornberg, K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43, 189–219 (2008).
Moss, B. in Fields Virology Vol. 5 (eds Knipe, D. M. & Howley, P. M.) 2906 (Lippincott-Raven, 2007).
Moss, B. Poxvirus cell entry: how many proteins does it take? Viruses 4, 688–707 (2012).
Hsiao, J. C., Chung, C. S. & Chang, W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73, 8750–8761 (1999).
Lin, C.-L., Chung, C.-S., Heine, H. G. & Chang, W. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74, 3353–3365 (2000).
Chiu, W.-L., Lin, C.-L., Yang, M.-H., Tzou, D.-L. M. & Chang, W. Vaccinia virus 4c (A26L) protein on intracellular mature virus binds to the extracellular cellular matrix laminin. J. Virol. 81, 2149–2157 (2007).
Chung, C. S., Hsiao, J. C., Chang, Y. S. & Chang, W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 72, 1577–1585 (1998).
Senkevich, T. G., Ward, B. M. & Moss, B. Vaccinia virus entry into cells is dependent on a virion surface protein encoded by the A28L gene. J. Virol. 78, 2357–2366 (2004).
Ulaeto, D., Grosenbach, D. & Hruby, D. E. The vaccinia virus 4c and A-type inclusion proteins are specific markers for the intracellular mature virus particle. J. Virol. 70, 3372–3377 (1996).
Law, M., Carter, G. C., Roberts, K. L., Hollinshead, M. & Smith, G. L. Ligand-induced and nonfusogenic dissolution of a viral membrane. Proc. Natl Acad. Sci. USA 103, 5989–5994 (2006).
Schmidt, F. I., Bleck, C. K. E., Helenius, A. & Mercer, J. Vaccinia extracellular virions enter cells by macropinocytosis and acid-activated membrane rupture. EMBO J. 30, 3647–3661 (2011).
Doms, R. W., Blumenthal, R. & Moss, B. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J. Virol. 64, 4884–4892 (1990).
Townsley, A. C., Weisberg, A. S., Wagenaar, T. R. & Moss, B. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J. Virol. 80, 8899–8908 (2006).
Gustafsson, M. G. L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87 (2000).
Gray, R. D. M. et al. VirusMapper: open-source nanoscale mapping of viral architecture through super-resolution microscopy. Sci. Rep. 6, 29132 (2016).
Gray, R. D. M., Mercer, J. & Henriques, R. Open-source single-particle analysis for super-resolution microscopy with VirusMapper. J. Vis. Exp. 122, e55471 (2017).
Unger, B. & Traktman, P. Vaccinia virus morphogenesis: A13 phosphoprotein is required for assembly of mature virions. J. Virol. 78, 8885–8901 (2004).
Senkevich, T. G., Ojeda, S., Townsley, A., Nelson, G. E. & Moss, B. Poxvirus multiprotein entry-fusion complex. Proc. Natl Acad. Sci. USA 102, 18572–18577 (2005).
Mirzakhanyan, Y. & Gershon, P. The Vaccinia virion: filling the gap between atomic and ultrastructure. PLoS Pathog. 15, e1007508 (2019).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Su, H.-P., Golden, J. W., Gittis, A. G., Hooper, J. W. & Garboczi, D. N. Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein. Virology 368, 331–341 (2007).
Levet, F. et al. SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat. Methods 12, 1065–1071 (2015).
Laliberte, J. P., Weisberg, A. S. & Moss, B. The membrane fusion step of vaccinia virus entry is cooperatively mediated by multiple viral proteins and host cell components. PLoS Pathog. 7, e1002446 (2011).
Rodriguez, J. F., Paez, E. & Esteban, M. A 14,000-Mr envelope protein of vaccinia virus is involved in cell fusion and forms covalently linked trimers. J. Virol. 61, 395–404 (1987).
Kochan, G., Escors, D., González, J. M., Casasnovas, J. M. & Esteban, M. Membrane cell fusion activity of the vaccinia virus A17-A27 protein complex. Cell. Microbiol. 10, 149–164 (2008).
Sanderson, C. M., Hollinshead, M. & Smith, G. L. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J. Gen. Virol. 81, 47–58 (2000).
Rodriguez, J. F. & Smith, G. L. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by golgi membrane and egress. Nucleic Acids Res. 18, 5347–5351 (1990).
Vázquez, M. I. & Esteban, M. Identification of functional domains in the 14-kilodalton envelope protein (A27L) of vaccinia virus. J. Virol. 73, 9098–9109 (1999).
Kiskowski, M. A., Hancock, J. F. & Kenworthy, A. K. On the use of Ripley’s K-function and its derivatives to analyze domain size. Biophys. J. 97, 1095–1103 (2009).
Schmidt, F. I., Kuhn, P., Robinson, T., Mercer, J. & Dittrich, P. S. Single-virus fusion experiments reveal proton influx into vaccinia virions and hemifusion lag times. Biophys. J. 105, 420–431 (2013).
Zawada, K. E., Okamoto, K. & Kasson, P. M. Influenza hemifusion phenotype depends on membrane context: differences in cell–cell and virus–cell fusion. J. Mol. Biol. 430, 594–601 (2018).
Chojnacki, J. et al. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338, 524–528 (2012).
Mercer, J. & Helenius, A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 320, 531–535 (2008).
Rizopoulos, Z. et al. Vaccinia virus infection requires maturation of macropinosomes. Traffic 16, 814–831 (2015).
Huang, C.-Y. et al. A novel cellular protein, VPEF, facilitates vaccinia virus penetration into HeLa cells through fluid phase endocytosis. J. Virol. 82, 7988–7999 (2008).
Vanderplasschen, A., Hollinshead, M. & Smith, G. L. Intracellular and extracellular vaccinia virions enter cells by different mechanisms. J. Gen. Virol. 79, 877–887 (1998).
Sandgren, K. J. et al. A differential role for macropinocytosis in mediating entry of the two forms of vaccinia virus into dendritic cells. PLoS Pathog. 6, e1000866 (2010).
Dales, S. The uptake and development of vaccinia virus in strain L cells followed with labeled viral deoxyribonucleic acid. J. Cell Biol. 18, 51–72 (1963).
Schmidt, F. I., Bleck, C. K. E. & Mercer, J. Poxvirus host cell entry. Curr. Opin. Virol. 2, 20–27 (2012).
Schmidt, F. I. et al. Vaccinia virus entry is followed by core activation and proteasome-mediated release of the immunomodulatory effector VH1 from lateral bodies. Cell Rep. 4, 464–476 (2013).
da Fonseca, F. G., Wolffe, E. J., Weisberg, A. & Moss, B. Effects of deletion or stringent repression of the H3L envelope gene on vaccinia virus replication. J. Virol. 74, 7518–7528 (2000).
Ojeda, S., Domi, A. & Moss, B. Vaccinia virus G9 protein is an essential component of the poxvirus entry-fusion complex. J. Virol. 80, 9822–9830 (2006).
Satheshkumar, P. S. & Moss, B. Characterization of a newly identified 35-amino-acid component of the vaccinia virus entry/fusion complex conserved in all chordopoxviruses. J. Virol. 83, 12822–12832 (2009).
Aldaz-Carroll, L. et al. Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R. J. Virol. 79, 6260–6271 (2005).
Szymborska, A. et al. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–658 (2013).
Heilemann, M. et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47, 6172–6176 (2008).
Nahidiazar, L., Agronskaia, A. V., Broertjes, J., van den Broek, B. & Jalink, K. Optimizing imaging conditions for demanding multi-color super resolution localization microscopy. PLoS ONE 11, e0158884 (2016).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Ovesný, M., Křížek, P., Borkovec, J., Švindrych, Z. & Hagen, G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
Laine, R. F. et al. NanoJ: a high-performance open-source super-resolution microscopy toolbox. J. Phys. D Appl. Phys. 52, 163001 (2019).
White, J. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89, 674–679 (1981).
Acknowledgements
We thank B. Moss and M. Esteban for providing the mutant viruses that were used in this study, and M. Turmaine (UCL Biosciences EM facility) and A. Weston (UCL School of Pharmacy—Electron Microscopy Unit) for use of their coater and SEM, respectively. We also acknowledge Eisenberg members J. C. Whitbeck, C. H. Foo and L. Aldaz-Carrol, of the poxvirus research group, for their help producing and purifying the VACV EFC antibodies. This work was funded by MRC Programme grant (MC_UU_00012/7; to J.M.), the European Research Council (649101, UbiProPox; to J.M.), the MRC (MR/K015826/1; to J.M. and R.H.), Biotechnology and Biological Sciences Research Council (BB/M022374/1, BB/P027431/1 and BB/R000697/1; to R.H.) and the Wellcome Trust (203276/Z/16/Z; to R.H.). R.D.M.G. is funded by the Engineering and Physical Sciences Research Council (EP/M506448/1). D.A. is a Marie Skłodowska-Curie fellow funded by the European Union (750673). C.B. is funded by the MRC LMCB PhD program.
Author information
Authors and Affiliations
Contributions
R.D.M.G., D.A., R.H. and J.M. conceived the project, designed the experiments and wrote the manuscript. R.D.M.G., D.A., C.B. and M.H. performed the experiments. G.H.C. helped to produce and purify all of the VACV EFC antibodies. I.J.W. and J.J.B. performed the EM. R.D.M.G., D.A. R.H and J.M analysed the data. R.D.M.G., D.A., R.H. and J.M. discussed the results and implications of the findings. All authors discussed the manuscript and provided comments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9, Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
Gray, R.D.M., Albrecht, D., Beerli, C. et al. Nanoscale polarization of the entry fusion complex of vaccinia virus drives efficient fusion. Nat Microbiol 4, 1636–1644 (2019). https://doi.org/10.1038/s41564-019-0488-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0488-4