Abstract
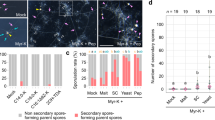
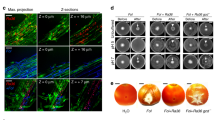
Arbuscular mycorrhizal (AM) fungi are obligate symbionts that depend on living host plants to complete their life cycle1,2. This feature, which leads to their unculturability in the absence of plants, strongly hinders basic research and agricultural application of AM fungi. However, at least one AM fungus can grow and develop fertile spores independently of a host plant in co-culture with the bacterium Paenibacillus validus3. The bacteria-derived substances are thought to act as stimulants or nutrients for fungal sporulation, but these molecules have not been identified. Here, we show that (S)-12-methyltetradecanoic acid4,5, a methyl branched-chain fatty acid isolated from bacterial cultures, stimulates the branching of hyphae germinated from mother spores and the formation of secondary spores in axenic culture of the AM fungus Rhizophagus irregularis. Extensive testing of fatty acids revealed that palmitoleic acid induces more secondary spores than the bacterial fatty acid in R. irregularis. These induced spores have the ability to infect host plant roots and to generate daughter spores. Our work shows that, in addition to a major source of organic carbon6,7,8,9, fatty acids act as stimulants to induce infection-competent secondary spores in the asymbiotic stage and could provide the key to developing the axenic production of AM inoculum.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bonfante, P. & Genre, A. Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1, 48 (2010).
Smith, S. E. & Read, D. J. Mycorrhizal Symbiosis (Academic Press, 2008).
Hildebrandt, U., Ouziad, F., Marner, F. J. & Bothe, H. The bacterium Paenibacillus validus stimulates growth of the arbuscular mycorrhizal fungus Glomus intraradices up to the formation of fertile spores. FEMS Microbiol. Lett. 254, 258–267 (2006).
Daane, L. L. et al. PAH-degradation by Paenibacillus spp. and description of Paenibacillus naphthalenovorans sp. nov., a naphthalene-degrading bacterium from the rhizosphere of salt marsh plants. Int. J. Syst. Evol. Microbiol. 52, 131–139 (2002).
Heyndrickx, M. et al. Paenibacillus (formerly Bacillus) gordonae (Pichinoty et al. 1986) Ash et al. 1994 is a later subjective synonym of Paenibacillus (formerly Bacillus) validus (Nakamura 1984) Ash et al. 1994: emended description of. Int. J. Syst. Bacteriol. 45, 661–669 (1995).
Bravo, A., Brands, M., Wewer, V., Dörmann, P. & Harrison, M. J. Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 214, 1631–1645 (2017).
Keymer, A. et al. Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 6, e29107 (2017).
Jiang, Y. et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175 (2017).
Luginbuehl, L. H. et al. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178 (2017).
Tisserant, E. et al. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl Acad. Sci. USA 110, 20117–20122 (2013).
Tang, N. et al. A survey of the gene repertoire of Gigaspora rosea unravels conserved features among glomeromycota for obligate biotrophy. Front. Microbiol. 7, 233 (2016).
Maeda, T. et al. Evidence of non-tandemly repeated rDNAs and their intragenomic heterogeneity in Rhizophagus irregularis. Commun. Biol. 1, 87 (2018).
Kitahara, T., Aono, S. & Mori, K. Synthesis of both the enantiomers of aseanostatin P5 (sarcinic acid), an inhibitor of myeloperoxidase release, and four diastereomers of aggreceride A, a platelet aggregation inhibitor. Biosci. Biotechnol. Biochem. 59, 78–82 (1995).
Hildebrandt, U., Janetta, K. & Bothe, H. Towards growth of arbuscular mycorrhizal fungi independent of a plant host towards growth of arbuscular mycorrhizal fungi Independent of a plant host. Appl. Environ. Microbiol. 68, 1919–1924 (2002).
Graham, J. H., Hodge, N. C. & Morton, J. B. Fatty acid methyl ester profiles for characterization of glomalean fungi and their endomycorrhizae. Appl. Environ. Microbiol. 61, 58–64 (1995).
Olsson, P. A. Signature fatty acids provide tools for determination of the distribution and interactions of mycorrhizal fungi in soil. FEMS Microbiol. Ecol. 29, 303–310 (1999).
Bentivenga, S. P. & Morton, J. B. Congruence of fatty acid methyl ester profiles and morphological characters of arbuscular mycorrhizal fungi in Gigasporaceae. Proc. Natl Acad. Sci. USA 93, 5659–5662 (1996).
Akiyama, K., Matsuzaki, K. & Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 (2005).
Besserer, A., Bécard, G., Jauneau, A., Roux, C. & Séjalon-Delmas, N. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 148, 402–413 (2008).
Besserer, A. et al. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4, e226 (2006).
Tsuzuki, S., Handa, Y., Takeda, N. & Kawaguchi, M. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol. Plant Microbe Interact. 29, 277–286 (2016).
Logi, C., Sbrana, C. & Giovannetti, M. Cellular events involved in survival of individual arbuscular mycorrhizal symbionts growing in the absence of the host. Appl. Environ. Microbiol. 64, 3473–3479 (1998).
Ryan, R. P. & Dow, J. M. Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 19, 145–152 (2011).
Zhu, S., Vivanco, J. M. & Manter, D. K. Nitrogen fertilizer rate affects root exudation, the rhizosphere microbiome and nitrogen-use-efficiency of maize. Appl. Soil Ecol. 107, 324–333 (2016).
Badri, D. V. & Vivanco, J. M. Regulation and function of root exudates. Plant Cell Environ. 32, 666–681 (2009).
Nadal, M. & Paszkowski, U. Polyphony in the rhizosphere: presymbiotic communication in arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 16, 473–479 (2013).
Frey-Klett, P., Garbaye, J. & Tarkka, M. The mycorrhiza helper bacteria revisited. New Phytol. 176, 22–36 (2007).
Bonfante, P. & Anca, I.-A. Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu. Rev. Microbiol. 63, 363–383 (2009).
Juge, C., Coughlan, A. P., Fortin, J. A. & Piché, Y. in Advances in Mycorrhizal Science and Technology (Khasa D. P. et al.) 39–50 (NRC Research Press, 2009).
Rosikiewicz, P., Bonvin, J. & Sanders, I. R. Cost-efficient production of in vitro Rhizophagus irregularis. Mycorrhiza 27, 477–486 (2017).
Maki, T., Nomachi, M., Yoshida, S. & Ezawa, T. Plant symbiotic microorganisms in acid sulfate soil: significance in the growth of pioneer plants. Plant Soil 310, 55–65 (2008).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Zhou, X., Lindsay, H. & Robinson, M. D. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. 42, e91 (2014).
Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Kikuchi, Y. et al. Polyphosphate accumulation is driven by transcriptome alterations that lead to near-synchronous and near-equivalent uptake of inorganic cations in an arbuscular mycorrhizal fungus. New Phytol. 204, 638–649 (2014).
Takeda, N. et al. Gibberellins interfere with symbiosis signaling and gene expression and alter colonization by arbuscular mycorrhizal fungi in Lotus japonicus. Plant Physiol. 167, 545–557 (2015).
Acknowledgements
We thank I. Nishida, H. Imai, T. Maeda, K. Ebine and M. Yamato for their critical suggestions, and Y. Ogawa, R. Oguchi, R. Okita, M. Ohashi, Y. Sugiura, S. Tanaka, A. Tokairin and Y. Yoshinori for their experimental support. We appreciate K. Yamaguchi, S. Shigenobu, Functional Genomics Facility and Data Integration and Analysis Facility and the NIBB Core Research Facilities for technical support. This work was supported by ACCEL (JPMJAC1403) from the Japan Science and Technology Agency; Grant-in-Aid for Scientific Research on Innovative Areas (22128006); Grant-in-Aid for Scientific Research (A) (15H01751) from the Japan Society for the Promotion of Science; and NIBB Collaborative Research Program (18-341).
Author information
Authors and Affiliations
Contributions
K.A. and H.H. designed the identification of the signal compounds and the bioassays of the compounds. I.T. identified the signal compounds and performed the bioassays of the compounds. H.K., K.S. and M.K. designed the RNA-seq analysis. H.K., Y.K., Y.H and T.E. performed the RNA-seq analysis. H.K. and K.S. performed the characterization of induced spores. H.K. analysed the data. H.K., K.S., M.K. and K.A. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 and Supplementary Tables 1–5.
Supplementary Table 4
Differentially expressed genes in response to palmitoleic acid (C16:1Δ9Z) and/or between hyphae and spores.
Supplementary Table 6
Genes co-regulated both by palmitoleic acid and intraradical mycelium.
Rights and permissions
About this article
Cite this article
Kameoka, H., Tsutsui, I., Saito, K. et al. Stimulation of asymbiotic sporulation in arbuscular mycorrhizal fungi by fatty acids. Nat Microbiol 4, 1654–1660 (2019). https://doi.org/10.1038/s41564-019-0485-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0485-7
This article is cited by
-
Spatial co-transcriptomics reveals discrete stages of the arbuscular mycorrhizal symbiosis
Nature Plants (2024)
-
Adding plant metabolites improve plant phosphorus uptake by altering the rhizosphere bacterial community structure
Plant and Soil (2024)
-
Control of arbuscule development by a transcriptional negative feedback loop in Medicago
Nature Communications (2023)
-
Disentangling arbuscular mycorrhizal fungi and bacteria at the soil-root interface
Mycorrhiza (2023)
-
Asymbiotic mass production of the arbuscular mycorrhizal fungus Rhizophagus clarus
Communications Biology (2022)