Abstract
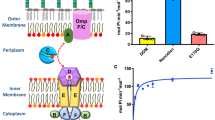
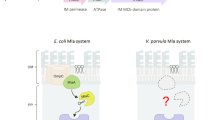
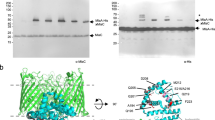
The Mla pathway is believed to be involved in maintaining the asymmetrical Gram-negative outer membrane via retrograde phospholipid transport. The pathway is composed of three components: the outer membrane MlaA–OmpC/F complex, a soluble periplasmic protein, MlaC, and the inner membrane ATPase, MlaFEDB complex. Here, we solve the crystal structure of MlaC in its phospholipid-free closed apo conformation, revealing a pivoting β-sheet mechanism that functions to open and close the phospholipid-binding pocket. Using the apo form of MlaC, we provide evidence that the inner-membrane MlaFEDB machinery exports phospholipids to MlaC in the periplasm. Furthermore, we confirm that the phospholipid export process occurs through the MlaD component of the MlaFEDB complex and that this process is independent of ATP. Our data provide evidence of an apparatus for lipid export away from the inner membrane and suggest that the Mla pathway may have a role in anterograde phospholipid transport.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The MlaC-apo X-ray structure has been deposited in the PDB with the accession number 6GKI. In addition, the data that support the findings of this study are available from the corresponding author on request.
References
Malinverni, J. C. & Silhavy, T. J. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl Acad. Sci. USA 106, 8009–8014 (2009).
Ruiz, N., Kahne, D. & Silhavy, T. J. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4, 57–66 (2006).
Kamio, Y. & Nikaido, H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase C and cyanogen bromide activated dextran in the external medium. Biochemistry 15, 2561–2570 (1976).
Nikaido, H. Restoring permeability barrier function to outer membrane. Chem. Biol. 12, 507–509 (2005).
Wu, T. et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 (2005).
Malinverni, J. C. et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61, 151–164 (2006).
Sklar, J. G. et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl Acad. Sci. USA 104, 6400–6405 (2007).
Knowles, T. J., Scott-Tucker, A., Overduin, M. & Henderson, I. R. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7, 206–214 (2009).
Ruiz, N., Gronenberg, L. S., Kahne, D. & Silhavy, T. J. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 105, 5537–5542 (2008).
Narita, S. & Tokuda, H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 583, 2160–2164 (2009).
Chng, S. S., Gronenberg, L. S. & Kahne, D. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49, 4565–4567 (2010).
Tran, A. X., Dong, C. & Whitfield, C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539 (2010).
Okuda, S. & Tokuda, H. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65, 239–259 (2011).
Grabowicz, M. & Silhavy, T. J. Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc. Natl Acad. Sci. USA 114, 4769–4774 (2017).
Jones, N. C. & Osborn, M. J. Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J. Biol. Chem. 252, 7405–7412 (1977).
Langley, K. E., Hawrot, E. & Kennedy, E. P. Membrane assembly: movement of phosphatidylserine between the cytoplasmic and outer membranes of Escherichia coli. J. Bacteriol. 152, 1033–1041 (1982).
Donohue-Rolfe, A. M. & Schaechter, M. Translocation of phospholipids from the inner to the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 77, 1867–1871 (1980).
Chong, Z. S., Woo, W. F. & Chng, S. S. Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol. Microbiol. 98, 1133–1146 (2015).
Thong, S. et al. Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. eLife 5, e19042 (2016).
Abellon-Ruiz, J. et al. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat. Microbiol. 2, 1616–1623 (2017).
Ekiert, D. C. et al. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169, 273–285 (2017).
Yeow, J. et al. The architecture of the OmpC-MlaA complex sheds light on the maintenance of outer membrane lipid asymmetry in Escherichia coli. J. Biol. Chem. 293, 11325–11340 (2018).
Powers, M. J. & Trent, M. S. Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc. Natl Acad. Sci. USA 115, E8518–E8527 (2018).
Kamischke, C. et al. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. eLife 8, e40171 (2019).
Suzuki, T. et al. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol. Microbiol. 11, 31–41 (1994).
Hong, M., Gleason, Y., Wyckoff, E. E. & Payne, S. M. Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect. Immun. 66, 4700–4710 (1998).
Cuccui, J. et al. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect. Immun. 75, 1186–1195 (2007).
Snijder, H. J. et al. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 401, 717–721 (1999).
Bishop, R. E. et al. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 19, 5071–5080 (2000).
Leney, A. C. & Heck, A. J. Native mass spectrometry: what is in the name? J. Am. Soc. Mass Spectrom. 28, 5–13 (2017).
Ercan, B., Low, W. Y., Liu, X. & Chng, S. S. Characterization of interactions and phospholipid transfer between substrate binding proteins of the OmpC-Mla system. Biochemistry 58, 114–119 (2018).
Giess, F., Friedrich, M. G., Heberle, J., Naumann, R. L. & Knoll, W. The protein-tethered lipid bilayer: a novel mimic of the biological membrane. Biophys. J. 87, 3213–3220 (2004).
Shen, H. H. et al. Reconstitution of a nanomachine driving the assembly of proteins into bacterial outer membranes. Nat. Commun. 5, 5078 (2014).
Norby, J. G. Coupled assay of Na+, K+ ATPase activity. Methods Enzym. 156, 116–119 (1988).
Henderson, J. C. et al. The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu. Rev. Microbiol. 70, 255–278 (2016).
Sutterlin, H. A. et al. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl Acad. Sci. USA 113, E1565–E1574 (2016).
Doerrler, W. T. & Raetz, C. R. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277, 36697–36705 (2002).
Eckford, P. D. & Sharom, F. J. The reconstituted Escherichia coli MsbA protein displays lipid flippase activity. Biochem. J. 429, 195–203 (2010).
Liu, C. E., Liu, P. Q. & Ames, G. F. Characterization of the adenosine triphosphatase activity of the periplasmic histidine permease, a traffic ATPase (ABC transporter). J. Biol. Chem. 272, 21883–21891 (1997).
Chen, J., Sharma, S., Quiocho, F. A. & Davidson, A. L. Trapping the transition state of an ATP-binding cassette transporter: evidence for a concerted mechanism of maltose transport. Proc. Natl Acad. Sci. USA 98, 1525–1530 (2001).
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Joosten, R. P. et al. PDB_REDO: automated re-refinement of X-ray structure models in the PDB. J. Appl. Crystallogr. 42, 376–384 (2009).
Kiianitsa, K., Solinger, J. A. & Heyer, W. D. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 321, 266–271 (2003).
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000).
Kay, L., Keifer, P. & Saarinen, T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114, 10663–10665 (1992).
Palmer III, A. G., Cavanagh, J., Byrd, R. A. & Rance, M. Sensitivity improvement in three-dimensional heteronuclear correlation NMR spectroscopy. J. Magn. Reson. 96, 416–424 (1992).
Schleucher, J. et al. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR 4, 301–306 (1994).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Goddard, T. D. & Kneller, D. G. SPARKY 3. University of California, San Francisco (2004).
Fiser, A. & Šali, A. in Methods in Enzymology, Vol. 374 (eds. Charles W. Carter, Jr & Robert, M. S.) 461–491 (Academic Press, 2003).
Li, D.-W. & Brüschweiler, R. NMR-based protein potentials. Angew. Chem. 122, 6930–6932 (2010).
Huang, J. & MacKerell, A. D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013).
Schmid, N. et al. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 40, 843–856 (2011).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Durell, S. R., Brooks, B. R. & Ben-Naim, A. Solvent-induced forces between two hydrophilic groups. J. Phys. Chem. 98, 2198–2202 (1994).
Neria, E., Fischer, S. & Karplus, M. Simulation of activation free energies in molecular systems. J. Chem. Phys. 105, 1902–1921 (1996).
Berendsen, H., Postma, J., van Gunsteren, W. & Hermans, J. in Intermolecular Forces (ed. Pullman, B.) 331–342 (D. Reidel Publishing Company, 1981).
Feenstra, K. A., Hess, B. & Berendsen, H. J. C. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J. Comput. Chem. 20, 786–798 (1999).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Nosé, S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 52, 255–268 (1984).
Hoover, W. G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A 31, 1695–1697 (1985).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Nosé, S. & Klein, M. L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 50, 1055–1076 (1983).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
Sugita, Y. & Okamoto, Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314, 141–151 (1999).
Bussi, G. Hamiltonian replica exchange in GROMACS: a flexible implementation. Mol. Phys. 112, 379–384 (2014).
Tribello, G. A., Bonomi, M., Branduardi, D., Camilloni, C. & Bussi, G. PLUMED 2: new feathers for an old bird. Comput. Phys. Commun. 185, 604–613 (2014).
Paramo, T., East, A., Garzón, D., Ulmschneider, M. B. & Bond, P. J. Efficient characterization of protein cavities within molecular simulation trajectories: trj_cavity. J. Chem. Theory Comput. 10, 2151–2164 (2014).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Sauerbrey, G. Z. The use of quartz oscillators for weighing thin layers and for microweighing. Z. für Phys. 155, 206–222 (1959).
Richter, R., Mukhopadhyay, A. & Brisson, A. Pathways of lipid vesicle deposition on solid surfaces: a combined QCM-D and AFM study. Biophys. J. 85, 3035–3047 (2003).
Reviakine, I., Johannsmann, D. & Richter, R. P. Hearing what you cannot see and visualizing what you hear: interpreting quartz crystal microbalance data from solvated interfaces. Anal. Chem. 83, 8838–8848 (2011).
Webster, J., Holt, S. & Dalgliesh, R. INTER the chemical interfaces reflectometer on target station 2 at ISIS. Phys. B 385–386, 1164–1166 (2006).
Born, M. & Wolf, E. Principles of Optics (Pergamon Press, 1970).
Clifton, L. A., Neylon, C. & Lakey, J. H. Examining protein-lipid complexes using neutron scattering. Methods Mol. Biol. 974, 119–150 (2013).
Clifton, L. A. et al. The effect of lipopolysaccharide core oligosaccharide size on the electrostatic binding of antimicrobial proteins to models of the Gram negative bacterial outer membrane. Langmuir 32, 3485–3494 (2016).
Clifton, L. A. et al. An accurate in vitro model of the E. coli envelope. Angew. Chem. Int. Ed. Engl. 54, 11952–11955 (2015).
Tamm, L. K. & McConnell, H. M. Supported phospholipid bilayers. Biophys. J. 47, 105–113 (1985).
Sivia, D. S. & Webster, J. R. P. The Bayesian approach to reflectivity data. Phys. B 248, 327–337 (1998).
Sivia, D. S. & Skilling, J. Data Analysis: a Bayesian Tutorial (Oxford Univ. Press, 2006).
Haario, H., Saksman, E. & Tamminen, J. An adaptive Metropolis algorithm. Bernoulli 7, 223–242 (2001).
Haario, H., Laine, M., Mira, A. & Saksman, E. DRAM: efficient adaptive MCMC. Stat. Comput. 16, 339–354 (2006).
Voss, N. R. & Gerstein, M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 38, W555–W562 (2010).
Acknowledgements
We thank D. Ekiert and G. Bhabha for their discussions and for providing the Mla plasmid constructs. We thank the HWB-NMR at the University of Birmingham for providing open access to their Wellcome Trust-funded NMR equipment. This research was supported by the BBSRC grant no. BB/P009840/1 (T.J.K. and G.W.H.) and Wellcome Trust grant no. 208400/Z/17/Z. T.J.P. acknowledges the use of the Iridis high-performance computing resources at the University of Southampton.
Author information
Authors and Affiliations
Contributions
G.W.H., S.C.L.H., C.S.L., P.S., C.H., T.J.P., P.J.W., A.C.L., D.G.W., M.J., V.S., I.T.C., C.H., G.L.I., J.A.B., Y.Y., M.J., D.H., I.R.H., L.A.C., A.L.L. and T.J.K. participated in the conception and design of the work. G.W.H., S.C.L.H., C.S.L., P.S., A.H.M., T.J.P., P.J.W., A.C.L., D.G.W., R.J.P., L.A.C., A.L.L. and T.J.K. participated in the data acquisition, analysis or interpretation of the work. G.W.H., S.C.L.H., C.S.L., P.S., T.J.P., P.J.W., A.C.L., D.G.W., L.A.C., A.L.L. and T.J.K. were involved in writing and editing the manuscript. All authors approved the final version submitted for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–28, Supplementary Tables 1–6, Supplementary Discussion and Supplementary References.
Rights and permissions
About this article
Cite this article
Hughes, G.W., Hall, S.C.L., Laxton, C.S. et al. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat Microbiol 4, 1692–1705 (2019). https://doi.org/10.1038/s41564-019-0481-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0481-y