Abstract
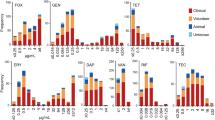
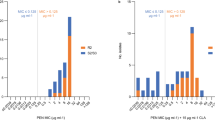
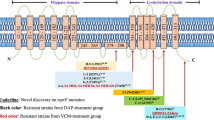
Antibiotic resistance in bacterial pathogens threatens the future of modern medicine. One such resistant pathogen is methicillin-resistant Staphylococcus aureus (MRSA), which is resistant to nearly all β-lactam antibiotics, limiting treatment options. Here, we show that a significant proportion of MRSA isolates from different lineages, including the epidemic USA300 lineage, are susceptible to penicillins when used in combination with β-lactamase inhibitors such as clavulanic acid. Susceptibility is mediated by a combination of two different mutations in the mecA promoter region that lowers mecA-encoded penicillin-binding protein 2a (PBP2a) expression, and in the majority of isolates by either one of two substitutions in PBP2a (E246G or M122I) that increase the affinity of PBP2a for penicillin in the presence of clavulanic acid. Treatment of S. aureus infections in wax moth and mouse models shows that penicillin/β-lactamase inhibitor susceptibility can be exploited as an effective therapeutic choice for ‘susceptible’ MRSA infection. Finally, we show that isolates with the PBP2a E246G substitution have a growth advantage in the presence of penicillin but the absence of clavulanic acid, which suggests that penicillin/β-lactamase susceptibility is an example of collateral sensitivity (resistance to one antibiotic increases sensitivity to another). Our findings suggest that widely available and currently disregarded antibiotics could be effective in a significant proportion of MRSA infections.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in the published article and its Supplementary Information files.
References
Worthington, R. J. & Melander, C. Overcoming resistance to β-lactam antibiotics. J. Org. Chem. 78, 4207–4213 (2013).
Hartman, B. J. & Tomasz, A. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158, 513–516 (1984).
Katayama, Y., Ito, T. & Hiramatsu, K. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44, 1549–1555 (2000).
Garcia-Alvarez, L. et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 11, 595–603 (2011).
Aedo, S. & Tomasz, A. Role of the stringent stress response in the antibiotic resistance phenotype of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 60, 2311–2317 (2016).
Rolinson, G. N., Stevens, S., Batchelor, F. R., Wood, J. C. & Chain, E. B. Bacteriological studies on a new penicillin—BRL.1241. Lancet 2, 564–567 (1960).
Reading, C. & Cole, M. Clavulanic acid: a β-lactamase-inhibiting β-lactam from Streptomyces clavuligerus. Antimicrob. Agents Chemother. 11, 852–857 (1977).
Chambers, H. F., Kartalija, M. & Sande, M. Ampicillin, sulbactam, and rifampin combination treatment of experimental methicillin-resistant Staphylococcus aureus endocarditis in rabbits. J. Infect. Dis. 171, 897–902 (1995).
Chambers, H. F., Sachdeva, M. & Kennedy, S. Binding affinity for penicillin-binding protein 2a correlates with in vivo activity of β-lactam antibiotics against methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 162, 705–710 (1990).
Washburn, R. G. & Durack, D. T. Efficacy of ampicillin plus a β-lactamase inhibitor (CP-45,899) in experimental endocarditis due to Staphylococcus aureus. J. Infect. Dis. 144, 237–243 (1981).
Cantoni, L., Wenger, A., Glauser, M. P. & Bille, J. Comparative efficacy of amoxicillin-clavulanate, cloxacillin, and vancomycin against methicillin-sensitive and methicillin-resistant Staphylococcus aureus endocarditis in rats. J. Infect. Dis. 159, 989–993 (1989).
Guignard, B., Entenza, J. M. & Moreillon, P. β-lactams against methicillin-resistant Staphylococcus aureus. Curr. Opin. Pharmacol. 5, 479–489 (2005).
Tan, C. M. et al. Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci. Transl. Med. 4, 126ra135 (2012).
Lee, S. H. et al. TarO-specific inhibitors of wall teichoic acid biosynthesis restore β-lactam efficacy against methicillin-resistant staphylococci. Sci. Transl. Med. 8, 329ra332 (2016).
Klitgaard, J. K., Skov, M. N., Kallipolitis, B. H. & Kolmos, H. J. Reversal of methicillin resistance in Staphylococcus aureus by thioridazine. J. Antimicrob. Chemother. 62, 1215–1221 (2008).
Ba, X. et al. Old drugs to treat resistant bugs: methicillin-resistant Staphylococcus aureus isolates with mecC are susceptible to a combination of penicillin and clavulanic acid. Antimicrob. Agents Chemother. 59, 7396–7404 (2015).
Weinstein, M. P. et al. M100 Performance Standards for Antimicrobial Susceptibility Testing 29th edn. (Clinical Laboratory Standards Institute, 2018).
Chambers, H. F. & Sachdeva, M. Binding of β-lactam antibiotics to penicillin-binding proteins in methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 161, 1170–1176 (1990).
Long, S. W. et al. PBP2a mutations causing high-level ceftaroline resistance in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 58, 6668–6674 (2014).
Otero, L. H. et al. How allosteric control of Staphylococcus aureus penicillin-binding protein 2a enables methicillin resistance and physiological function. Proc. Natl Acad. Sci. USA 110, 16808–16813 (2013).
Harkins, C. P. et al. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 18, 130 (2017).
Gill, S. R. et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187, 2426–2438 (2005).
Loeffler, A. et al. First isolation of MRSA ST398 from UK animals: a new challenge for infection control teams? J. Hosp. Infect. 72, 269–271 (2009).
Paterson, G. K. et al. Incidence and characterisation of methicillin-resistant Staphylococcus aureus (MRSA) from nasal colonisation in participants attending a cattle veterinary conference in the UK. PLoS ONE 8, e68463 (2013).
Everitt, B. S. An introduction to finite mixture distributions. Stat. Methods Med. Res. 5, 107–127 (1996).
Hernandez-Garcia, J. et al. Patterns of antimicrobial resistance in Streptococcus suis isolates from pigs with or without streptococcal disease in England between 2009 and 2014. Vet. Microbiol. 207, 117–124 (2017).
Chen, F. J., Wang, C. H., Chen, C. Y., Hsu, Y. C. & Wang, K. T. Role of the mecA gene in oxacillin resistance in a Staphylococcus aureus clinical strain with a pvl-positive ST59 genetic background. Antimicrob. Agents Chemother. 58, 1047–1054 (2014).
Ender, M., McCallum, N. & Berger-Bachi, B. Impact of mecA promoter mutations on mecA expression and β-lactam resistance levels. Int. J. Med. Microbiol. 298, 607–617 (2008).
DANMAP (Statums Serum Institute, 2018); https://www.danmap.org/
Coll, F. et al. Longitudinal genomic surveillance of MRSA reveals extensive transmission in hospitals and the community. Sci. Transl. Med. 9, eaak9745 (2017).
Toleman, M. S. et al. Systematic surveillance detects multiple silent introductions and household transmission of methicillin-resistant Staphylococcus aureus USA300 in the east of England. J. Infect. Dis. 214, 447–453 (2016).
Jamrozy, D. M. et al. Pan-genomic perspective on the evolution of the Staphylococcus aureus USA300 epidemic. Microb. Genom. 2, e000058 (2016).
Uhlemann, A. C. et al. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc. Natl Acad. Sci. USA 111, 6738–6743 (2014).
Hartman, B. J. & Tomasz, A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 29, 85–92 (1986).
Larsen, A. R. et al. Two distinct clones of methicillin-resistant Staphylococcus aureus (MRSA) with the same USA300 pulsed-field gel electrophoresis profile: a potential pitfall for identification of USA300 community-associated MRSA. J. Clin. Microbiol. 47, 3765–3768 (2009).
Collins, J. et al. Offsetting virulence and antibiotic resistance costs by MRSA. ISME J. 4, 577–584 (2010).
Rudkin, J. K. et al. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J. Infect. Dis. 205, 798–806 (2012).
Pozzi, C. et al. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 8, e1002626 (2012).
Pal, C., Papp, B. & Lazar, V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 23, 401–407 (2015).
BSAC Methods for Antimicrobial Susceptibility Testing Version 14 (British Society for Antimicrobial Chemotherapy, 2015).
Ersoy, S. C. et al. Correcting a fundamental flaw in the paradigm for antimicrobial susceptibility testing. EBioMedicine 20, 173–181 (2017).
Martin, C. et al. Comparison of concentrations of two doses of clavulanic acid (200 and 400 milligrams) administered with amoxicillin (2,000 milligrams) in tissues of patients undergoing colorectal surgery. Antimicrob. Agents Chemother. 39, 94–98 (1995).
Kahlmeter, G. The 2014 Garrod Lecture: EUCAST—are we heading towards international agreement? J. Antimicrob. Chemother. 70, 2427–2439 (2015).
Nicolas, M. H., Kitzis, M. D. & Karim, A. Stérilisation par 2 grammes d’Augmentin® des urines infectées à Staphylococcus aureus résistant à la méticilline. Med. Mal. Infect. 23, 82–94 (1993).
Franciolli, M., Bille, J., Glauser, M. P. & Moreillon, P. β-lactam resistance mechanisms of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 163, 514–523 (1991).
Andreoni, M., Raillard, P., Concia, E. & Wang, Y. Sulbactam/ampicillin in the treatment of skin and soft-tissue infections due of methicillin-resistance staphylococci. A pilot study. Curr. Ther. Res. Clin. Exp. 50, 386–395 (1991).
Nigo, M. et al. Ceftaroline-resistant, daptomycin-tolerant, and heterogeneous vancomycin-intermediate methicillin-resistant Staphylococcus aureus causing infective endocarditis. Antimicrob. Agents Chemother. 61, e01235-16 (2017).
Steenbergen, J. N., Mohr, J. F. & Thorne, G. M. Effects of daptomycin in combination with other antimicrobial agents: a review of in vitro and animal model studies. J. Antimicrob. Chemother. 64, 1130–1138 (2009).
Dilworth, T. J. et al. β-lactams enhance vancomycin activity against methicillin-resistant Staphylococcus aureus bacteremia compared to vancomycin alone. Antimicrob. Agents Chemother. 58, 102–109 (2014).
Reed, P. et al. Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog. 11, e1004891 (2015).
Andrews, J. M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48, 5–16 (2001).
Monk, I. R., Shah, I., Xu, M., Tan, M.-W. & Foster, T. J. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3, 11 (2012).
Corrigan, R. M. & Foster, T. J. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61, 126–129 (2009).
Grilo, I. R., Ludovice, A. M., Tomasz, A., de Lencastre, H. & Sobral, R. G. The glucosaminidase domain of Atl—the major Staphylococcus aureus autolysin—has DNA-binding activity. MicrobiologyOpen 3, 247–256 (2014).
Zhao, G., Meier, T. I., Kahl, S. D., Gee, K. R. & Blaszczak, L. C. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43, 1124–1128 (1999).
Kim, C. et al. Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the β-lactam-resistant phenotype. J. Biol. Chem. 287, 36854–36863 (2012).
Bouley, R. et al. Discovery of antibiotic (E)-3-(3-carboxyphenyI)-2-(4-cyanostyryl)quinazolin-4(3H)-one. J. Am. Chem. Soc. 137, 1738–1741 (2015).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Desbois, A. P. & Coote, P. J. Wax moth larva (Galleria mellonella): an in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 66, 1785–1790 (2011).
Laabei, M. et al. Evolutionary trade-offs underlie the multi-faceted virulence of Staphylococcus aureus. PLoS Biol. 13, e1002229 (2015).
Ziebuhr, W. et al. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65, 890–896 (1997).
Page, A. J. et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb. Genom. 2, e000083 (2016).
Gladman, S. & Seemann, T. Velvet Optimiser: automate your Velvet assemblies (GitHub, 2008); https://github.com/tseemann/VelvetOptimiser
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D. & Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2011).
Boetzer, M. & Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 13, R56 (2012).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Gouy, M., Guindon, S. & Gascuel, O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010).
Ambler, R. P. et al. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276, 269–270 (1991).
Voladri, R. K. & Kernodle, D. S. Characterization of a chromosomal gene encoding type B β-lactamase in phage group II isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 42, 3163–3168 (1998).
Diep, B. A. et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739 (2006).
Croucher, N. J. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15 (2015).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Lim, D. & Strynadka, N. C. Structural basis for the β lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 9, 870–876 (2002).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank L. R. Hansen Kildevang and A. Medina for testing of the Danish isolates. We thank É. Brouillette and F. Malouin for providing isolates. This work was supported by Medical Research Council partnership grants (G1001787/1 and MR/N002660/1) held between the Department of Veterinary Medicine and School of Clinical Medicine at the University of Cambridge, the Moredun Research Institute and the Wellcome Sanger Institute. This publication presents independent research supported by the Health Innovation Challenge Fund (WT098600 and HICF-T5-342)—a parallel funding partnership between the Department of Health and Wellcome Trust. The views expressed in this publication are those of the author(s) and not necessarily those of the Department of Health or Wellcome Trust. E.M.H. is supported by a UK Research and Innovation Fellowship (MR/S00291X/1). F.C. is supported by the Wellcome Trust (201344/Z/16/Z). X.B. is supported by a UK–China AMR Partnership Grant (MR/P007201/1).
Author information
Authors and Affiliations
Contributions
E.M.H., X.B., S.J.P. and M.A.H. designed the study. X.B. performed the mecA deletion and complementation, expression analysis, and bocillin assays. X.B., B.B., N.G. and K.L.B. did the antimicrobial susceptibility testing. H.C. and R.C.M. ran the biofilm and toxicity assays. J.L. and A.R.L. determined the antimicrobial susceptibility testing of Danish isolates. O.R. determined the ECOFF. A.L. analysed the structure of PBP2a. E.M.H., X.B. and C.V.L. performed the infection and treatment experiments. I.R.G. and R.S. ran the bocillin binding assays. E.M.H., F.C., S.R. and D.J. performed the bioinformatics analysis of whole-genome sequence data. A.-C.U. and F.D.L. collected the USA300 isolates. N.G. wrote the bioinformatics scripts. C.U.K., G.K.P., M.T.G.H. and J.P. analysed and interpreted the data. E.M.H. coordinated the study and wrote the manuscript. S.J.P. and M.A.H. supervised and managed the study. All authors read, contributed to and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
C.U.K. is a consultant for the World Health Organization Regional Office for Europe, QuantuMDx Group and Foundation for Innovative New Diagnostics, which involves work for Cepheid, Hain Lifescience and the World Health Organization. C.U.K. is an advisor to GenoScreen. The European Society of Mycobacteriology awarded C.U.K. the Gertrud Meissner Award, which is sponsored by Hain Lifescience. The Bill and Melinda Gates Foundation, Janssen Pharmaceutica and PerkinElmer covered C.U.K.’s travel and accommodation to enable presentations at meetings. The Global Alliance for TB Drug Development and Otsuka Novel Products have supplied C.U.K. with antibiotics for in vitro research. C.U.K. has collaborated with Illumina on a number of scientific projects. S.J.P. and J.P. are consultants to Next Gen Diagnostics. S.J.P. is a consultant to Specific Technologies. All other authors have no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7, Tables 2 and 6–8, Supplementary Discussion, Supplementary References, and footnotes for Supplementary Tables 1 and 3–5.
Supplementary Table 1
Key features of 110 MRSA isolates screened for β-lactam resistance.
Supplementary Table 3
Key features of 298 MRSA isolates screened for penicillin–clavulanic acid susceptibility.
Supplementary Table 4
Key features of Danish clinical isolates screened for penicillin–clavulanic acid susceptibility.
Supplementary Table 5
Relevant characteristics of CC8 and USA300 included in Fig. 4b.
Rights and permissions
About this article
Cite this article
Harrison, E.M., Ba, X., Coll, F. et al. Genomic identification of cryptic susceptibility to penicillins and β-lactamase inhibitors in methicillin-resistant Staphylococcus aureus. Nat Microbiol 4, 1680–1691 (2019). https://doi.org/10.1038/s41564-019-0471-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0471-0
This article is cited by
-
Multimodal nanoimmunotherapy engages neutrophils to eliminate Staphylococcus aureus infections
Nature Nanotechnology (2024)
-
Transcription tuned by S-nitrosylation underlies a mechanism for Staphylococcus aureus to circumvent vancomycin killing
Nature Communications (2023)
-
Cryptic susceptibility to penicillin/β-lactamase inhibitor combinations in emerging multidrug-resistant, hospital-adapted Staphylococcus epidermidis lineages
Nature Communications (2023)
-
Amino acid (acyl carrier protein) ligase-associated biosynthetic gene clusters reveal unexplored biosynthetic potential
Molecular Genetics and Genomics (2023)
-
Phenotypic and genomic characteristics of oxacillin-susceptible mecA-positive Staphylococcus aureus, rapid selection of high-level resistance to beta-lactams
European Journal of Clinical Microbiology & Infectious Diseases (2023)