Abstract
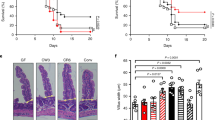
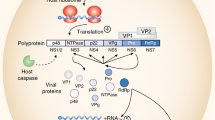
Commensal microbes profoundly impact host immunity to enteric viral infections1. We have shown that the bacterial microbiota and host antiviral cytokine interferon-λ (IFN-λ) determine the persistence of murine norovirus in the gut2,3. However, the effects of the virome in modulating enteric infections remain unexplored. Here, we report that murine astrovirus can complement primary immunodeficiency to protect against murine norovirus and rotavirus infections. Protection against infection was horizontally transferable between immunocompromised mouse strains by co-housing and fecal transplantation. Furthermore, protection against enteric pathogens corresponded with the presence of a specific strain of murine astrovirus in the gut, and this complementation of immunodeficiency required IFN-λ signalling in gut epithelial cells. Our study demonstrates that elements of the virome can protect against enteric pathogens in an immunodeficient host.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Code availability
The code used for constructing the phylogenetic tree is available at GitHub (https://github.com/RachelRodgers/STL5-phylogenetics).
Data availability
The data from this study are available in the main paper and Supplementary Information. Sequencing data have been uploaded to the European Nucleotide Archive with accession number PRJEB31115.
References
Pfeiffer, J. K. & Virgin, H. W. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 351, aad5872 (2016).
Baldridge, M. T. et al. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science 347, 266–269 (2015).
Nice, T. J. et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347, 269–273 (2015).
Netea, M. G. et al. Trained immunity: a program of innate immune memory in health and disease. Science 352, aaf1098 (2016).
Virgin, H. W. The virome in mammalian physiology and disease. Cell 157, 142–150 (2014).
Rosshart, S. P. et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171, 1015–1028 (2017).
Beura, L. K. et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516 (2016).
Kaufmann, E. et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell 172, 176–190 (2018).
Kleinnijenhuis, J. et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl Acad. Sci. USA 109, 17537–17542 (2012).
Arts, R. J. W. et al. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe 23, 89–100 (2018).
Barton, E. S. et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447, 326–329 (2007).
Sun, J. C., Beilke, J. N. & Lanier, L. L. Adaptive immune features of natural killer cells. Nature 457, 557–561 (2009).
Chudnovskiy, A. et al. Host–protozoan interactions protect from mucosal infections through activation of the inflammasome. Cell 167, 444–456 (2016).
Nice, T. J., Strong, D. W., McCune, B. T., Pohl, C. S. & Virgin, H. W. A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. J. Virol. 87, 327–334 (2013).
Thackray, L. B. et al. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 81, 10460–10473 (2007).
Mombaerts, P. et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992).
Shinkai, Y. et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68, 855–867 (1992).
Cao, X. et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity 2, 223–238 (1995).
Chachu, K. A., LoBue, A. D., Strong, D. W., Baric, R. S. & Virgin, H. W. Immune mechanisms responsible for vaccination against and clearance of mucosal and lymphatic norovirus infection. PLoS Pathog. 4, e1000236 (2008).
Chachu, K. A. et al. Antibody is critical for the clearance of murine norovirus infection. J. Virol. 82, 6610–6617 (2008).
Zhang, B. et al. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science 346, 861–865 (2014).
Uchiyama, R., Chassaing, B., Zhang, B. & Gewirtz, A. T. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis. 210, 171–182 (2014).
Galvez, E. J. C., Iljazovic, A., Gronow, A., Flavell, R. & Strowig, T. Shaping of intestinal microbiota in Nlrp6- and Rag2-deficient mice depends on community structure. Cell Rep. 21, 3914–3926 (2017).
Handley, S. A. et al. SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe 19, 323–335 (2016).
Norman, J. M., Handley, S. A. & Virgin, H. W. Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology 146, 1459–1469 (2014).
Zhao, G. et al. VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology 503, 21–30 (2017).
Yokoyama, C. C. et al. Adaptive immunity restricts replication of novel murine astroviruses. J. Virol. 86, 12262–12270 (2012).
Lee, S. et al. Norovirus cell tropism is determined by combinatorial action of a viral non-structural protein and host cytokine. Cell Host Microbe 22, 449–459 (2017).
Baldridge, M. T. et al. Expression of Ifnlr1 on intestinal epithelial cells is critical to the antiviral effects of IFN-lambda against norovirus and reovirus. J. Virol. 91, e02079-16 (2017).
Shultz, L. D., Brehm, M. A., Garcia-Martinez, J. V. & Greiner, D. L. Humanized mice for immune system investigation: progress, promise and challenges. Nat. Rev. Immunol. 12, 786–798 (2012).
Strong, D. W., Thackray, L. B., Smith, T. J. & Virgin, H. W. Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J. Virol. 86, 2950–2958 (2012).
Orchard, R. C. et al. Discovery of a proteinaceous cellular receptor for a norovirus. Science 353, 933–936 (2016).
Feng, N., Burns, J. W., Bracy, L. & Greenberg, H. B. Comparison of mucosal and systemic humoral immune responses and subsequent protection in mice orally inoculated with a homologous or a heterologous rotavirus. J. Virol. 68, 7766–7773 (1994).
Moon, C. et al. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature 521, 90–93 (2015).
Baert, L. et al. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appli. Environ. Microbiol. 74, 543–546 (2008).
Baym, M. et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS ONE 10, e0128036 (2015).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108, 4516–4522 (2011).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
McDonald, D. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618 (2012).
Vazquez-Baeza, Y., Pirrung, M., Gonzalez, A. & Knight, R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2, 16 (2013).
Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J. & Sayers, E. W. GenBank. Nucleic Acids Res. 44, D67–D72 (2016).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Acknowledgements
We acknowledge all members of the Baldridge laboratory for helpful discussions. We also thank J. Hoisington-Lopez for assistance with sequencing, D. Kreamalmeyer for animal care and breeding, the Washington University Pulmonary Morphology core facility for assistance with histology, and J. White and the Washington University Central Gnotobiotic Facility for assistance with germ-free mice. We are grateful to the Estes laboratory for providing murine rotavirus. H.I. was supported by the Children’s Discovery Institute of Washington University and a St Louis Children’s Hospital Postdoctoral Research grant (MI-F-2018-712). S.L. was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (NRF-2016R1A6A3A03012352). A.O. was funded by the Pediatric Infectious Diseases Society/St Jude Children’s Research Hospital Fellowship Program in Basic Research and NIH training grant T32AI106688. C.C.Y. was supported by NIH training grant T32AI007163. S.S.-C. was supported by ALSAC and NIH grant R03 AI126101-01. J.J.M. was supported by NIH grant K08 AR07091. M.T.B. was supported by NIH grants K22 AI127846, R01 AI127552, R01 AI139314 and R01 AI141478, Digestive Diseases Research Core Centers P30 DK052574, the Pew Biomedical Scholars Program and the Global Probiotics Council’s Young Investigator Grant for Probiotics Research.
Author information
Authors and Affiliations
Contributions
H.I., S.L., T.A., M.S., S.T.P., M.L., T.-C.L., C.C.Y. and M.T.B. performed the experiments. H.I., S.L., T.A., A.O., G.Z., M.S., S.T.P., M.L., T.C.L., C.C.Y., B.S., R.R., S.S.-C., J.J.M. and M.T.B analysed the results. H.I., S.L., S.S.-C., J.J.M and M.T.B designed the project. H.I., S.L. and M.T.B. wrote the manuscript. All authors read and edited the manuscript. All reagents are available from M.T.B. under a material transfer agreement with Washington University.
Corresponding author
Ethics declarations
Competing interests
C.C.Y. and G.Z. hold a patent for detection of murine astrovirus. The authors declare no additional conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1-8.
Rights and permissions
About this article
Cite this article
Ingle, H., Lee, S., Ai, T. et al. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-λ. Nat Microbiol 4, 1120–1128 (2019). https://doi.org/10.1038/s41564-019-0416-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0416-7
This article is cited by
-
Exploring the Human Virome: Composition, Dynamics, and Implications for Health and Disease
Current Microbiology (2024)
-
Chromosome folding and prophage activation reveal specific genomic architecture for intestinal bacteria
Microbiome (2023)
-
Moniezia benedeni infection enhances neuromedin U (NMU) expression in sheep (Ovis aries) small intestine
BMC Veterinary Research (2022)
-
Intestinal immunoregulation: lessons from human mendelian diseases
Mucosal Immunology (2021)
-
The human virome: assembly, composition and host interactions
Nature Reviews Microbiology (2021)