Abstract
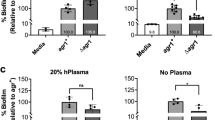
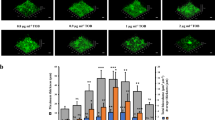
Social interactions play an increasingly recognized key role in bacterial physiology1. One of the best studied is quorum sensing (QS), a mechanism by which bacteria sense and respond to the status of cell density2. While QS is generally deemed crucial for bacterial survival, QS-dysfunctional mutants frequently arise in in vitro culture. This has been explained by the fitness cost an individual mutant, a ‘quorum cheater’, saves at the expense of the community3. QS mutants are also often isolated from biofilm-associated infections, including cystic fibrosis lung infection4, as well as medical device infection and associated bacteraemia5,6,7. However, despite the frequently proposed use of QS blockers to control virulence8, the mechanisms underlying QS dysfunctionality during infection have remained poorly understood. Here, we show that in the major human pathogen Staphylococcus aureus, quorum cheaters arise exclusively in biofilm infection, while in non-biofilm-associated infection there is a high selective pressure to maintain QS control. We demonstrate that this infection-type dependence is due to QS-dysfunctional bacteria having a significant survival advantage in biofilm infection because they form dense and enlarged biofilms that provide resistance to phagocyte attacks. Our results link the benefit of QS-dysfunctional mutants in vivo to biofilm-mediated immune evasion, thus to mechanisms that are specific to the in vivo setting. Our findings explain why QS mutants are frequently isolated from biofilm-associated infections and provide guidance for the therapeutic application of QS blockers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information files) or available from the corresponding author upon request.
References
Asfahl, K. L. & Schuster, M. Social interactions in bacterial cell–cell signaling. FEMS Microbiol. Rev. 41, 92–107 (2017).
Miller, M. B. & Bassler, B. L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 (2001).
Dandekar, A. A., Chugani, S. & Greenberg, E. P. Bacterial quorum sensing and metabolic incentives to cooperate. Science 338, 264–266 (2012).
Bjarnsholt, T. et al. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS ONE 5, e10115 (2010).
Vuong, C., Kocianova, S., Yao, Y., Carmody, A. B. & Otto, M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190, 1498–1505 (2004).
Traber, K. E. et al. agr function in clinical Staphylococcus aureus isolates. Microbiology 154, 2265–2274 (2008).
Fowler, V. G. et al. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190, 1140–1149 (2004).
Dickey, S. W., Cheung, G. Y. C. & Otto, M. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 16, 457–471 (2017).
Rumbaugh, K. P. et al. Quorum sensing and the social evolution of bacterial virulence. Curr. Biol. 19, 341–345 (2009).
Pollitt, E. J., West, S. A., Crusz, S. A., Burton-Chellew, M. N. & Diggle, S. P. Cooperation, quorum sensing, and evolution of virulence in Staphylococcus aureus. Infect. Immun. 82, 1045–1051 (2014).
Gordon, R. J. & Lowy, F. D. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46, S350–S359 (2008).
Le, K. Y. & Otto, M. Quorum-sensing regulation in staphylococci—an overview. Front. Microbiol. 6, 1174 (2015).
Otto, M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 64, 175–188 (2013).
Periasamy, S. et al. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl Acad. Sci. USA 109, 1281–1286 (2012).
Peschel, A. & Otto, M. Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 11, 667–673 (2013).
Wang, R. et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514 (2007).
Painter, K. L., Krishna, A., Wigneshweraraj, S. & Edwards, A. M. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia?. Trends Microbiol. 22, 676–685 (2014).
Collins, J. et al. Offsetting virulence and antibiotic resistance costs by MRSA. ISME J. 4, 577–584 (2010).
Paulander, W. et al. Antibiotic-mediated selection of quorum-sensing-negative Staphylococcus aureus. mBio 3, e00459–00412 (2013).
Vuong, C., Saenz, H. L., Gotz, F. & Otto, M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182, 1688–1693 (2000).
Shopsin, B. et al. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 202, 1593–1599 (2010).
Vuong, C., Kocianova, S., Yu, J., Kadurugamuwa, J. L. & Otto, M. Development of real-time in vivo imaging of device-related Staphylococcus epidermidis infection in mice and influence of animal immune status on susceptibility to infection. J. Infect. Dis. 198, 258–261 (2008).
Li, M. et al. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl Acad. Sci. USA 106, 5883–5888 (2009).
Carrel, M., Perencevich, E. N. & David, M. Z. USA300 methicillin-resistant Staphylococcus aureus, United States, 2000–2013. Emerg. Infect. Dis. 21, 1973–1980 (2015).
Kourbatova, E. V. et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA 300 clone as a cause of health care-associated infections among patients with prosthetic joint infections. Am. J. Infect. Control 33, 385–391 (2005).
Foster, T. J. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3, 948–958 (2005).
Cheung, G. Y., Wang, R., Khan, B. A., Sturdevant, D. E. & Otto, M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79, 1927–1935 (2011).
Butterfield, J. M. et al. Predictors of agr dysfunction in methicillin-resistant Staphylococcus aureus (MRSA) isolates among patients with MRSA bloodstream infections. Antimicrob. Agents Chemother. 55, 5433–5437 (2011).
Nowakowska, J., Landmann, R. & Khanna, N. Foreign body infection models to study host–pathogen response and antimicrobial tolerance of bacterial biofilm. Antibiotics (Basel) 3, 378–397 (2014).
Flannagan, R. S., Heit, B. & Heinrichs, D. E. Antimicrobial mechanisms of macrophages and the immune evasion strategies of staphylococcus aureus. Pathogens 4, 826–868 (2015).
Joo, H. S., Cheung, G. Y. & Otto, M. Antimicrobial activity of community-associated methicillin-resistant Staphylococcus aureus is caused by phenol-soluble modulin derivatives. J. Biol. Chem. 286, 8933–8940 (2011).
Diep, B. A., Carleton, H. A., Chang, R. F., Sensabaugh, G. F. & Perdreau-Remington, F. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 193, 1495–1503 (2006).
Somerville, G. A. et al. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J. Bacteriol. 184, 1430–1437 (2002).
Voyich, J. M. et al. Is Panton–Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194, 1761–1770 (2006).
Wang, R. et al. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest. 121, 238–248 (2011).
Cox, P. J., Phillips, B. J. & Thomas, P. The enzymatic basis of the selective action of cyclophosphamide. Cancer Res. 35, 3755–3761 (1975).
Acknowledgements
This study was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), U.S. National Institutes of Health (NIH) (project no. 1 ZIA AI000904) to M.O., The National Natural Science Foundation of China (grant no. 81501804) to L.H. and the General Program of Shanghai Municipal Commission of Health and Family Planning (grant no. 20154Y0014) to L.H.
Author information
Authors and Affiliations
Contributions
L.H., K.Y.L. and R.L.H. performed confocal microscopy. B.A.K. performed in vitro quorum cheating assays. L.H., T.H.N., G.Y.C.C., J.S.B., R.L.H. and Y.Z. performed animal experiments. L.H., T.H.N. and R.L.H. performed leukocyte killing assays. L.H. performed all other experiments. L.H., K.Y.L., G.Y.C.C., J.K., J.S.B. and M.O. analysed the data. M.L and M.O. designed and supervised experiments. M.O. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, and Supplementary Figures 1–4.
Rights and permissions
About this article
Cite this article
He, L., Le, K.Y., Khan, B.A. et al. Resistance to leukocytes ties benefits of quorum sensing dysfunctionality to biofilm infection. Nat Microbiol 4, 1114–1119 (2019). https://doi.org/10.1038/s41564-019-0413-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0413-x
This article is cited by
-
The role of Staphylococcus aureus quorum sensing in cutaneous and systemic infections
Inflammation and Regeneration (2024)
-
Single-cell transcriptome reveals Staphylococcus aureus modulating fibroblast differentiation in the bone-implant interface
Molecular Medicine (2023)
-
Antibiotic treatment can exacerbate biofilm-associated infection by promoting quorum cheater development
npj Biofilms and Microbiomes (2023)
-
Probiotic disruption of quorum sensing reduces virulence and increases cefoxitin sensitivity in methicillin-resistant Staphylococcus aureus
Scientific Reports (2023)
-
Nanomaterial-based strategies in antimicrobial applications: Progress and perspectives
Nano Research (2021)