Abstract
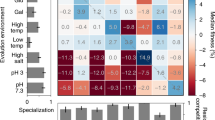
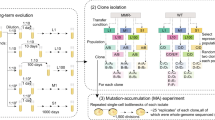
How does environmental complexity affect the evolution of single genes? Here, we measured the effects of a set of Bacillus subtilis glutamate dehydrogenase mutants across 19 different environments—from phenotypically homogeneous single-cell populations in liquid media to heterogeneous biofilms, plant roots and soil populations. The effects of individual gene mutations on organismal fitness were highly reproducible in liquid cultures. However, 84% of the tested alleles showed opposing fitness effects under different growth conditions (sign environmental pleiotropy). In colony biofilms and soil samples, different alleles dominated in parallel replica experiments. Accordingly, we found that in these heterogeneous cell populations the fate of mutations was dictated by a combination of selection and drift. The latter relates to programmed prophage excisions that occurred during biofilm development. Overall, for each condition, a wide range of glutamate dehydrogenase mutations persisted and sometimes fixated as a result of the combined action of selection, pleiotropy and chance. However, over longer periods and in multiple environments, nearly all of this diversity would be lost—across all the environments and conditions that we tested, the wild type was the fittest allele.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author on request.
References
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Koo, B. M. et al. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 4, 291–305 (2017).
Pache, R. A., Madan, M. M. & Aloy, P. Exploiting gene deletion fitness effects in yeast to understand the modular architecture of protein complexes under different growth conditions. BMC Syst. Biol. 3, 74 (2009).
Winzeler, E. A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999).
Civelek, M. & Lusis, A. J. Systems genetics approaches to understand complex traits. Nat. Rev. Genet. 15, 34–48 (2014).
Steinberg, B. & Ostermeier, M. Environmental changes bridge evolutionary valleys. Sci. Adv. 2, e1500921 (2016).
Wang, Z., Liao, B.-Y. & Zhang, J. Genomic patterns of pleiotropy and the evolution of complexity. Proc. Natl Acad. Sci. USA 107, 18034–18039 (2010).
Fusco, D., Gralka, M., Kayser, J., Anderson, A. & Hallatschek, O. Excess of mutational jackpot events in expanding populations revealed by spatial Luria-Delbrück experiments. Nat. Commun. 7, 12760 (2016).
Kimura, M. Genetic variability maintained in a finite population due to mutational production of neutral and nearly neutral isoalleles. Genet. Res. 89, 341–363 (2008).
Smith, N. H., Gordon, S. V., de la Rua-Domenech, R., Clifton-Hadley, R. S. & Hewinson, R. G. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat. Rev. Microbiol. 4, 670–681 (2006).
Nei, M. Selectionism and neutralism in molecular evolution. Mol. Biol. Evol. 22, 2318–2342 (2005).
Steenackers, H. P., Parijs, I., Foster, K. R. & Vanderleyden, J. Experimental evolution in biofilm populations. FEMS Microbiol. Rev. 40, 373–397 (2016).
Boucher, J. I., Bolon, D. N. A. & Tawfik, D. S. Quantifying and understanding the fitness effects of protein mutations: laboratory versus nature. Protein Sci. 25, 1219–1226 (2016).
De Visser, J. A. G. M. & Krug, J. Empirical fitness landscapes and the predictability of evolution. Nat. Rev. Genet. 15, 480–490 (2014).
Fowler, D. M. & Fields, S. Deep mutational scanning: a new style of protein science. Nat. Methods 11, 801–807 (2014).
Steinberg, B. & Ostermeier, M. Shifting fitness and epistatic landscapes reflect trade-offs along an evolutionary pathway. J. Mol. Biol. 428, 2730–2743 (2016).
Dandage, R. et al. Differential strengths of molecular determinants guide environment specific mutational fates. PLoS Genet. 14, e1007419 (2018).
Earl, A. M., Losick, R. & Kolter, R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16, 269–275 (2008).
Branda, S. S., González-Pastor, J. E., Ben-Yehuda, S., Losick, R. & Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl Acad. Sci. USA 98, 11621–11626 (2001).
Belitsky, B. R. & Sonenshein, A. L. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 180, 6298–6305 (1998).
Gunka, K. & Commichau, F. M. Control of glutamate homeostasis in Bacillus subtilis: a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation. Mol. Microbiol. 85, 213–224 (2012).
Liu, J. et al. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 523, 550–554 (2015).
Noda‐Garcia, L., Romero Romero, M. L., Longo, L. M., Kolodkin‐Gal, I. & Tawfik, D. S. Bacilli glutamate dehydrogenases diverged via coevolution of transcription and enzyme regulation. EMBO Rep. 18, 1139–1149 (2017).
De Jong, L. et al. In-culture cross-linking of bacterial cells reveals large-scale dynamic protein-protein interactions at the peptide level. J. Proteome Res. 16, 2457–2471 (2017).
Stannek, L. et al. Evidence for synergistic control of glutamate biosynthesis by glutamate dehydrogenases and glutamate in Bacillus subtilis. Environ. Microbiol. 17, 3379–3390 (2015).
Mondal, S., Yakhnin, A. V., Sebastian, A., Albert, I. & Babitzke, P. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat. Microbiol. 1, 15007 (2016).
Besharova, O., Suchanek, V. M., Hartmann, R., Drescher, K. & Sourjik, V. Diversification of gene expression during formation of static submerged biofilms by Escherichia coli. Front. Microbiol. 7, 1568 (2016).
Vlamakis, H., Chai, Y., Beauregard, P., Losick, R. & Kolter, R. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168 (2013).
Mavor, D. et al. Determination of ubiquitin fitness landscapes under different chemical stresses in a classroom setting. eLife 5, e15802 (2016).
Driffield, K., Miller, K., Bostock, J. M., O’Neill, A. J. & Chopra, I. Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 61, 1053–1056 (2008).
Hallatschek, O., Hersen, P., Ramanathan, S. & Nelson, D. R. Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl Acad. Sci. USA 104, 19926–19930 (2007).
Hölscher, T. et al. Monitoring spatial segregation in surface colonizing microbial populations. J. Vis. Exp. 116, e54752 (2016).
Van Gestel, J., Weissing, F. J., Kuipers, O. P. & Kovács, Á. T. Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 8, 2069–2079 (2014).
Kunkel, B., Losick, R. & Stragier, P. The Bacillus subtilis gene for the developmental transcription factor sigma K is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 4, 525–535 (1990).
Nicolas, P. et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106 (2012).
Westers, H. et al. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 20, 2076–2090 (2003).
Eichenberger, P. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2, e328 (2004).
Abe, K. Developmentally-regulated excision of the SPβ prophage reconstitutes a gene required for spore envelope maturation in Bacillus subtilis. PLoS Genet. 10, e1004636 (2014).
Sanchez-Vizuete, P. et al. Identification of ypqP as a new Bacillus subtilis biofilm determinant that mediates the protection of Staphylococcus aureus against antimicrobial agents in mixed-species communities. Appl. Environ. Microbiol. 81, 109–118 (2015).
Pérez-Osorio, A. C., Williamson, K. S. & Franklin, M. J. Heterogeneous rpoS and rhlR mRNA levels and 16S rRNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms, sampled by laser capture microdissection. J. Bacteriol. 192, 2991–3000 (2010).
Vesper, O. et al. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147, 147–157 (2011).
Miton, C. M. & Tokuriki, N. How mutational epistasis impairs predictability in protein evolution and design. Protein Sci. 25, 1260–1272 (2016).
Breen, M. S., Kemena, C., Vlasov, P. K., Notredame, C. & Kondrashov, F. A. Epistasis as the primary factor in molecular evolution. Nature 490, 535–538 (2012).
Konkol, M. A., Blair, K. M. & Kearns, D. B. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J. Bacteriol. 195, 4085–4093 (2013).
Yi, Y., de Jong, A., Frenzel, E. & Kuipers, O. P. Comparative transcriptomics of Bacillus mycoides root exudates reveals different genetic adaptation of endophytic and soil isolates. Front. Microbiol. 8, 1487 (2017).
Deatherage, D. E. & Barrick, J. E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014).
Acknowledgements
L.N.-G. was supported by CONACYT grant no. 203740 and the Martin Kushner Fellowship at the Weizmann Institute of Science. D.S.T. is the Nella and Leon Benoziyo Professor of Biochemistry. Financial support from the Kahn Centre for Systems Biology at the Weizmann Institute of Science is gratefully acknowledged. We thank R. Milo, S. Fleishman, Z. Livneh and F. Kondrashov for their support and critical advice and E. Segev and A. de Visser for their critical and insightful comments on the manuscript. We appreciate the help of M. Hershko with script development for data processing and of Y. Bar-On and S. Gleizer with the analysis of genomic sequences. We are grateful to R. Rotkopf from Weizmann Life Sciences Core Facilities for his guidance on the statistical analysis. We are thankful for the services provided by the Crown Genomics Institute of the Nancy and Stephen Grand Israel National Centre for Personalized Medicine, Weizmann Institute of Science.
Author information
Authors and Affiliations
Contributions
L.N.-G. and D.S.T. designed the experiments and wrote the manuscript. L.N.-G., D.D. and D.S.T. analysed the data. L.N.-G. performed all experiments, except the selection of the soil colonization that was performed in collaboration with E.K. and A.A. D.D. and A.E. wrote the scripts used for the data analysis and visualization. E.P. applied the machine learning classification.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–12, Supplementary Tables 1–10 and legends for Supplementary Datasets.
Supplementary Dataset 1
Illumina raw counts for all alleles measured in each experiment. All codons that encode for the wild-type amino acid were summed under ‘WT’ and are shown in bold numbers. Sheet no. 1 shows the Illumina reads raw counts for the selection conditions that were initiated from Initial Mix 1 and 2: liquid, pellicle biofilms, colony biofilms, spores and germinated spores. Sheet no. 2 shows the Illumina reads raw counts for the conditions initiated from Initial mix 3: bulk soil. For each condition, three replica experiments were performed except for the bulk soil condition, where five replica experiments were performed.
Supplementary Dataset 2
FC values for all alleles and experiments. The codes assigning the individual conditions and experiments are described in Supplementary Tables 2 and 3. Sheet no. 1 shows the FC values for all conditions initiated from Initial Mix 1 and 2: liquid, pellicle biofilms, colony biofilms, spores and germinated spores. Sheet no. 2 shows the FC values for all conditions initiated from Initial Mix 3: bulk soil.
Supplementary Dataset 3
Analysis of SNPs and mobile elements in the sequenced genomes of various populations. The position in the genome, the mutations, their frequency and the mutated gene/protein (if applicable) are shown. Sheet no. 1 provides a description of all sheets in this file.
Rights and permissions
About this article
Cite this article
Noda-García, L., Davidi, D., Korenblum, E. et al. Chance and pleiotropy dominate genetic diversity in complex bacterial environments. Nat Microbiol 4, 1221–1230 (2019). https://doi.org/10.1038/s41564-019-0412-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0412-y
This article is cited by
-
Environmental selection and epistasis in an empirical phenotype–environment–fitness landscape
Nature Ecology & Evolution (2022)
-
A complex struggle for direction
Nature Chemical Biology (2022)
-
The adaptive landscape of a metallo-enzyme is shaped by environment-dependent epistasis
Nature Communications (2021)
-
Bacillus subtilis biofilm formation and social interactions
Nature Reviews Microbiology (2021)
-
MaveDB: an open-source platform to distribute and interpret data from multiplexed assays of variant effect
Genome Biology (2019)