Abstract
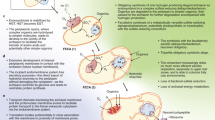
The origin of eukaryotes represents an unresolved puzzle in evolutionary biology. Current research suggests that eukaryotes evolved from a merger between a host of archaeal descent and an alphaproteobacterial endosymbiont. The discovery of the Asgard archaea, a proposed archaeal superphylum that includes Lokiarchaeota, Thorarchaeota, Odinarchaeota and Heimdallarchaeota suggested to comprise the closest archaeal relatives of eukaryotes, has helped to elucidate the identity of the putative archaeal host. Whereas Lokiarchaeota are assumed to employ a hydrogen-dependent metabolism, little is known about the metabolic potential of other members of the Asgard superphylum. We infer the central metabolic pathways of Asgard archaea using comparative genomics and phylogenetics to be able to refine current models for the origin of eukaryotes. Our analyses indicate that Thorarchaeota and Lokiarchaeota encode proteins necessary for carbon fixation via the Wood–Ljungdahl pathway and for obtaining reducing equivalents from organic substrates. By contrast, Heimdallarchaeum LC2 and LC3 genomes encode enzymes potentially enabling the oxidation of organic substrates using nitrate or oxygen as electron acceptors. The gene repertoire of Heimdallarchaeum AB125 and Odinarchaeum indicates that these organisms can ferment organic substrates and conserve energy by coupling ferredoxin reoxidation to respiratory proton reduction. Altogether, our genome analyses suggest that Asgard representatives are primarily organoheterotrophs with variable capacity for hydrogen consumption and production. On this basis, we propose the ‘reverse flow model’, an updated symbiogenetic model for the origin of eukaryotes that involves electron or hydrogen flow from an organoheterotrophic archaeal host to a bacterial symbiont.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Code availability
The small custom scripts used for genome annotation and phylogenetic analyses are made available on figshare and can be accessed at the following link: https://figshare.com/s/5f153d1dcacadd3b3ed6.
Data availability
The genomes of the herein analysed Asgard archaea have been made publicly available on NCBI previously2,4. Detailed annotations of the metabolic repertoire are provided in Supplementary Tables 1–3 accompanying this paper. Raw data files are made available via figshare under the following link: https://figshare.com/s/5f153d1dcacadd3b3ed6.
References
Hug, L. A. et al. A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016).
Spang, A. et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015).
Seitz, K. W., Lazar, C. S., Hinrichs, K. U., Teske, A. P. & Baker, B. J. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J. 10, 1696–1705 (2016).
Zaremba-Niedzwiedzka, K. et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017).
Spang, A., Caceres, E. F. & Ettema, T. J. G. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science 357, eaaf3883 (2017).
Lopez-Garcia, P. & Moreira, D. Open questions on the origin of eukaryotes. Trends Ecol. Evol. 30, 697–708 (2015).
Guy, L., Saw, J. H. & Ettema, T. J. The archaeal legacy of eukaryotes: a phylogenomic perspective. Cold Spring Harb. Perspect. Biol. 6, a016022 (2014).
Martin, W. F., Garg, S. & Zimorski, V. Endosymbiotic theories for eukaryote origin. Phil. Trans. R. Soc. B 370, 20140330 (2015).
Da Cunha, V., Gaia, M., Gadelle, D., Nasir, A. & Forterre, P. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 13, e1006810 (2017).
Spang, A. et al. Asgard archaea are the closest prokaryotic relatives of eukaryotes. PLoS Genet. 14, e1007080 (2018).
Narrowe, A. B. et al. Complex evolutionary history of translation elongation factor 2 and diphthamide biosynthesis in archaea and parabasalids. Genome Biol. Evol. 10, 2380–2393 (2018).
Klinger, C. M., Spang, A., Dacks, J. B. & Ettema, T. J. G. Tracing the archaeal origins of eukaryotic membrane-trafficking system building blocks. Mol. Biol. Evol. 33, 1528–1541 (2016).
Sousa, F. L., Neukirchen, S., Allen, J. F., Lane, N. & Martin, W. F. Lokiarchaeon is hydrogen dependent. Nat. Microbiol. 4, 16034 (2016).
Martin, W. F., Tielens, A. G. M., Mentel, M., Garg, S. G. & Gould, S. B. The physiology of phagocytosis in the context of mitochondrial origin. Microbiol. Mol. Biol. Rev. 81, e00008-17 (2017).
Zachar, I., Szilagyi, A., Szamado, S. & Szathmary, E. Farming the mitochondrial ancestor as a model of endosymbiotic establishment by natural selection. Proc. Natl Acad. Sci. USA 115, E1504–E1510 (2018).
Speijer, D. Alternating terminal electron-acceptors at the basis of symbiogenesis: how oxygen ignited eukaryotic evolution. Bioessays 39, 1600174 (2017).
Koonin, E. V. Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: eukaryogenesis just made easier? Phil. Trans. R. Soc. B 370, 20140333 (2015).
Williams, T. A., Foster, P. G., Cox, C. J. & Embley, T. M. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504, 231–236 (2013).
Lopez-Garcia, P., Eme, L. & Moreira, D. Symbiosis in eukaryotic evolution. J. Theor. Biol. 434, 20–33 (2017).
Ragsdale, S. W. & Pierce, E. Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784, 1873–1898 (2008).
Schuchmann, K. & Muller, V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821 (2014).
Adam, P. S., Borrel, G., Brochier-Armanet, C. & Gribaldo, S. The growing tree of Archaea: new perspectives on their diversity, evolution and ecology. ISME J. 11, 2407–2425 (2017).
Liu, Y. et al. Comparative genomic inference suggests mixotrophic lifestyle for Thorarchaeota. ISME J. 12, 1021–1031 (2018).
Wagner, A. et al. Mechanisms of gene flow in archaea. Nat. Rev. Microbiol. 15, 492–501 (2017).
Buckel, W. & Thauer, R. K. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113 (2013).
Bryant, F. O. & Adams, M. W. Characterization of hydrogenase from the hyperthermophilic archaebacterium, Pyrococcus furiosus. J. Biol. Chem. 264, 5070–5079 (1989).
Schuchmann, K. & Muller, V. Energetics and application of heterotrophy in acetogenic bacteria. Appl. Environ. Microbiol. 82, 4056–4069 (2016).
Stams, A. J. & Plugge, C. M. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7, 568–577 (2009).
Greening, C. et al. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10, 761–777 (2016).
Yu, H. et al. Structure of an ancient respiratory system. Cell 173, 1636–1649.e16 (2018).
Schut, G. J., Boyd, E. S., Peters, J. W. & Adams, M. W. The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol. Rev. 37, 182–203 (2013).
Tully, B. J., Graham, E. D. & Heidelberg, J. F. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data 5, 170203 (2018).
Adam, P. S., Borrel, G. & Gribaldo, S. Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc. Natl Acad. Sci. USA 115, E1166–E1173 (2018).
Kono, T. et al. A RuBisCO-mediated carbon metabolic pathway in methanogenic archaea. Nat. Commun. 8, 14007 (2017).
Lang, B. F., Gray, M. W. & Burger, G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33, 351–397 (1999).
Arshad, A. et al. A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front. Microbiol. 6, 1423 (2015).
Williams, T. A. et al. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc. Natl Acad. Sci. USA 114, E4602–E4611 (2017).
Zachar, I. & Szathmary, E. Breath-giving cooperation: critical review of origin of mitochondria hypotheses: major unanswered questions point to the importance of early ecology. Biol. Direct 12, 19 (2017).
Moreira, D. & Lopez-Garcia, P. Symbiosis between methanogenic archaea and delta-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J. Mol. Evol. 47, 517–530 (1998).
López-García, P. & Moreira, D. Selective forces for the origin of the eukaryotic nucleus. Bioessays 28, 525–533 (2006).
Martin, W. & Muller, M. The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41 (1998).
Sieber, J. R., McInerney, M. J. & Gunsalus, R. P. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu. Rev. Microbiol. 66, 429–452 (2012).
McGlynn, S. E., Chadwick, G. L., Kempes, C. P. & Orphan, V. J. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535 (2015).
Wegener, G., Krukenberg, V., Riedel, D., Tegetmeyer, H. E. & Boetius, A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526, 587–590 (2015).
Knittel, K. & Boetius, A. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334 (2009).
Seitz, K. W. et al. New Asgard archaea capable of anaerobic hydrocarbon cycling. Preprint at https://www.biorxiv.org/content/10.1101/527697v2 (2019).
Laso-Perez, R. et al. Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature 539, 396–401 (2016).
Martijn, J., Vosseberg, J., Guy, L., Offre, P. & Ettema, T. J. G. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557, 101–105 (2018).
Leger, M. M., Eme, L., Stairs, C. W. & Roger, A. J. Demystifying eukaryote lateral gene transfer (response to Martin 2017 DOI: 10.1002/bies.201700115). Bioessays 40, e1700242 (2018).
Stairs, C. W. et al. Microbial eukaryotes have adapted to hypoxia by horizontal acquisitions of a gene involved in rhodoquinone biosynthesis. eLife 7, e34292 (2018).
Norlund, K. L. et al. Microbial architecture of environmental sulfur processes: a novel syntrophic sulfur-metabolizing consortia. Environ. Sci. Technol. 43, 8781–8786 (2009).
Bose, A., Gardel, E. J., Vidoudez, C., Parra, E. A. & Girguis, P. R. Electron uptake by iron-oxidizing phototrophic bacteria. Nat. Commun. 5, 3391 (2014).
Eme, L., Spang, A., Lombard, J., Stairs, C. W. & Ettema, T. J. G. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 15, 711–723 (2017).
Ettema, T. J. Evolution: mitochondria in the second act. Nature 531, 39–40 (2016).
Caforio, A. et al. Converting Escherichia coli into an archaebacterium with a hybrid heterochiral membrane. Proc. Natl Acad. Sci. USA 115, 3704–3709 (2018).
Martin, W. et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165 (1998).
Pittis, A. A. & Gabaldon, T. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 531, 101–104 (2016).
Roger, A. J., Munoz-Gomez, S. A. & Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192 (2017).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Archaeal Clusters of Orthologous Genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life (Basel) 5, 818–840 (2015).
Saier, M. H. Jr, Tran, C. V. & Barabote, R. D. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34, D181–D186 (2006).
Sondergaard, D., Pedersen, C. N. & Greening, C. HydDB: a web tool for hydrogenase classification and analysis. Sci. Rep. 6, 34212 (2016).
Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017).
Yin, Y. et al. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451 (2012).
Rawlings, N. D., Barrett, A. J. & Finn, R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44, D343–D350 (2016).
Lenfant, N. et al. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 41, D423–D429 (2013).
Yu, N. Y. et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 (2010).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Criscuolo, A. & Gribaldo, S. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Wang, H. C., Minh, B. Q., Susko, E. & Roger, A. J. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst. Biol. 67, 216–235 (2018).
Kamikawa, R. et al. Parallel re-modeling of EF-1α function: divergent EF-1α genes co-occur with EFL genes in diverse distantly related eukaryotes. BMC Evol. Biol. 13, 131 (2013).
Lartillot, N., Rodrigue, N., Stubbs, D. & Richer, J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611–615 (2013).
Susko, E. & Roger, A. J. On reduced amino acid alphabets for phylogenetic inference. Mol. Biol. Evol. 24, 2139–2150 (2007).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Capella-Gutierrez, S. et al. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Minh, B. Q., Nguyen, M. A. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Anantharaman, K. et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7, 13219 (2016).
Wrighton, K. C. et al. RuBisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J. 10, 2702–2714 (2016).
Swigonova, Z., Mohsen, A. W. & Vockley, J. Acyl-CoA dehydrogenases: dynamic history of protein family evolution. J. Mol. Evol. 69, 176–193 (2009).
Dibrova, D. V., Galperin, M. Y. & Mulkidjanian, A. Y. Phylogenomic reconstruction of archaeal fatty acid metabolism. Environ. Microbiol. 16, 907–918 (2014).
Hug, L. A. et al. Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Phil. Trans. R. Soc. B 368, 20120322 (2013).
Jugder, B. E., Ertan, H., Lee, M., Manefield, M. & Marquis, C. P. Reductive dehalogenases come of age in biological destruction of organohalides. Trends Biotechnol. 33, 595–610 (2015).
Neumann, A., Wohlfarth, G. & Diekert, G. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 180, 4140–4145 (1998).
Vignais, P. M. & Billoud, B. Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272 (2007).
Rochette, N. C., Brochier-Armanet, C. & Gouy, M. Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes. Mol. Biol. Evol. 31, 832–845 (2014).
Acknowledgements
This work was supported by grants from the European Research Council (ERC starting grant 310039-PUZZLE_CELL to T.J.G.E.), the Swedish Foundation for Strategic Research (SSF-FFL5 to T.J.G.E.), the Swedish Research Council (VR grant 2015-04959 to T.J.G.E. and VR starting grant 2016-03559 to A.S.), the NWO-I Foundation of the Netherlands Organisation for Scientific Research (WISE fellowship to A.S.), the European Commission (Marie Curie IEF European grants 625521 to A.S. and 704263 to L.E.), the Wenner-Gren Foundations in Stockholm (2016-0072 to J.L.), the European Molecular Biology Organization (EMBO long-term fellowship ALTF-997–2015 to C.W.S.), the Natural Sciences and Engineering Research Council of Canada (C.W.S), the Australian Research Council (DE170100310 and DP180101762 to C.G.) and the National Science Foundation (DEB: Systematics and Biodiversity Sciences; award number 1737298 to B.J.B.). We thank K. Zaremba-Niedzwiedzka and J. Saw for reconstruction of some of these genomes and helpful discussions. We also acknowledge S. L. Jørgensen, the chief scientist R. B. Pedersen, the scientific party and the entire crew on board the Norwegian research vessel G.O. Sars during the summer 2010 expedition, which allowed us access to samples from Loki’s Castle. Finally, we thank P. Offre for discussions on metabolic inferences.
Author information
Authors and Affiliations
Contributions
A.S. and T.J.G.E. conceived the study. A.S., C.W.S., E.F.C., J.L., C.G., B.J.B. and N.D. analysed the genomic data. A.S., C.W.S. and L.E. performed the phylogenetic analyses. A.S. and T.J.G.E. wrote the manuscript with input from all authors. A.S., C.W.S. and N.D. wrote the Supplementary Information. All documents were edited and approved by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Text, Supplementary References, legends for Supplementary Tables, Supplementary Figures 1–18 and Supplementary Files 1–3.
Supplementary Tables 1–4
Overview of the presence/absence of discussed enzymes in Asgard lineages; annotations for proteins, which serve as candidate enzymes potentially involved in the various metabolic pathways discussed throughout this manuscript; automatic annotation of all genes; carbohydrate active enzymes, peptidases, esterases and information on extracellular localization.
Supplementary Table 5
Annotation of beta-oxidation genes encoded by Asgard genomes per protein family/phylogeny.
Rights and permissions
About this article
Cite this article
Spang, A., Stairs, C.W., Dombrowski, N. et al. Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nat Microbiol 4, 1138–1148 (2019). https://doi.org/10.1038/s41564-019-0406-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0406-9
This article is cited by
-
Evolution of optimal growth temperature in Asgard archaea inferred from the temperature dependence of GDP binding to EF-1A
Nature Communications (2024)
-
ATP synthase evolution on a cross-braced dated tree of life
Nature Communications (2023)
-
Actin cytoskeleton and complex cell architecture in an Asgard archaeon
Nature (2023)
-
Eco-evolutionary modelling of microbial syntrophy indicates the robustness of cross-feeding over cross-facilitation
Scientific Reports (2023)
-
Energetics and evolution of anaerobic microbial eukaryotes
Nature Microbiology (2023)