Abstract
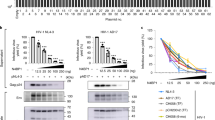
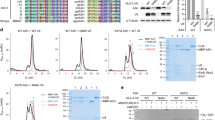
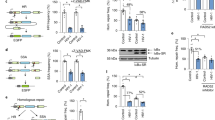
Type 1 interferon suppresses viral replication by upregulating the expression of interferon-stimulated genes with diverse antiviral properties1. The replication of human immunodeficiency virus type 1 (HIV-1) is naturally inhibited by interferon, with the steps between viral entry and chromosomal integration of viral DNA being notably susceptible2,3,4,5. The interferon-stimulated gene myxovirus resistance 2 has been defined as an effective postentry inhibitor of HIV-1, but is only partially responsible for interferon’s suppressive effect6,7,8. Using small interfering RNA-based library screening in interferon-α-treated cells, we sought to characterize further interferon-stimulated genes that target the pre-integration phases of HIV-1 infection, and identified human tripartite-containing motif 5α (TRIM5α) as a potent anti-HIV-1 restriction factor. Human TRIM5α, in contrast with many nonhuman orthologues, has not generally been ascribed substantial HIV-1 inhibitory function, a finding attributed to ineffective recognition of cytoplasmic viral capsids by TRIM5α2,9,10. Here, we demonstrate that interferon-α-mediated stimulation of the immunoproteasome, a proteasome isoform mainly present in immune cells and distinguished from the constitutive proteasome by virtue of its different catalytic β-subunits, as well as the proteasome activator 28 regulatory complex11,12,13, and the associated accelerated turnover of TRIM5α underpin the reprogramming of human TRIM5α for effective capsid-dependent inhibition of HIV-1 DNA synthesis and infection. These observations identify a mechanism for regulating human TRIM5α antiviral function in human cells and rationalize how TRIM5α participates in the immune control of HIV-1 infection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011).
Malim, M. H. & Bieniasz, P. D. HIV restriction factors and mechanisms of evasion. Cold Spring Harb. Perspect. Med. 2, a006940 (2012).
Goujon, C. & Malim, M. H. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J. Virol. 84, 9254–9266 (2010).
Doyle, T., Goujon, C. & Malim, M. H. HIV-1 and interferons: who’s interfering with whom? Nat. Rev. Microbiol. 13, 403–413 (2015).
Cheney, K. M. & McKnight, A. Interferon-alpha mediates restriction of human immunodeficiency virus type-1 replication in primary human macrophages at an early stage of replication. PLoS ONE 5, e13521 (2010).
Goujon, C. et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502, 559–562 (2013).
Kane, M. et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502, 563–566 (2013).
Liu, Z. et al. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14, 398–410 (2013).
Stremlau, M. et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 (2004).
Sayah, D. M., Sokolskaja, E., Berthoux, L. & Luban, J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430, 569–573 (2004).
Akiyama, K. et al. Replacement of proteasome subunits X and Y by LMP7 and LMP2 induced by interferon-gamma for acquirement of the functional diversity responsible for antigen processing. FEBS Lett. 343, 85–88 (1994).
Ma, C. P., Slaughter, C. A. & DeMartino, G. N. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J. Biol. Chem. 267, 10515–10523 (1992).
Groettrup, M., Kirk, C. J. & Basler, M. Proteasomes in immune cells: more than peptide producers? Nat. Rev. Immunol. 10, 73–78 (2010).
Wagner, J. M. et al. General model for retroviral capsid pattern recognition by trim5 proteins. J. Virol. 92, e01563-17 (2018).
Pham, Q. T., Bouchard, A., Grutter, M. G. & Berthoux, L. Generation of human TRIM5alpha mutants with high HIV-1 restriction activity. Gene Ther. 17, 859–871 (2010).
Yap, M. W., Nisole, S. & Stoye, J. P. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15, 73–78 (2005).
Fletcher, A. J. et al. TRIM5alpha requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription. EMBO J. 34, 2078–2095 (2015).
Campbell, E. M. et al. TRIM5alpha-mediated ubiquitin chain conjugation is required for inhibition of hiv-1 reverse transcription and capsid destabilization. J. Virol. 90, 1849–1857 (2016).
Diaz-Griffero, F. et al. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virol. 349, 300–315 (2006).
Sawyer, S. L., Wu, L. I., Akey, J. M., Emerman, M. & Malik, H. S. High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5alpha in humans. Curr. Biol. 16, 95–100 (2006).
Carthagena, L. et al. Human TRIM gene expression in response to interferons. PLoS ONE 4, e4894 (2009).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Preckel, T. et al. Impaired immunoproteasome assembly and immune responses in PA28-/- mice. Science 286, 2162–2165 (1999).
Kloetzel, P. M. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2, 179–187 (2001).
Muchamuel, T. et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med. 15, 781–787 (2009).
Granier, C. et al. Pressure from TRIM5alpha contributes to control of HIV-1 replication by individuals expressing protective HLA-B alleles. J. Virol. 87, 10368–10380 (2013).
Ribeiro, C. M. et al. Receptor usage dictates HIV-1 restriction by human TRIM5alpha in dendritic cell subsets. Nature 540, 448–452 (2016).
Sewram, S. et al. Human TRIM5alpha expression levels and reduced susceptibility to HIV-1 infection. J. Infect. Dis. 199, 1657–1663 (2009).
Celerino da Silva, R. et al. TRIM5 gene polymorphisms in HIV-1-infected patients and healthy controls from Northeastern Brazil. Immunol. Res. 64, 1237–1242 (2016).
vanManen, D. et al. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 4, e18 (2008).
Schindler, M. et al. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 77, 10548–10556 (2003).
Naldini, L. et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267 (1996).
Cordeil, S. et al. Evidence for a different susceptibility of primate lentiviruses to type I interferons. J. Virol. 87, 2587–2596 (2013).
Ochsenbauer, C. et al. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 86, 2715–2728 (2012).
Parrish, N. F. et al. Phenotypic properties of transmitted founder HIV-1. Proc. Natl Acad. Sci. USA 110, 6626–6633 (2013).
Dicks, M. D. et al. Oligomerization requirements for MX2-mediated suppression of HIV-1 infection. J. Virol. 90, 22–32 (2015).
Treier, M., Staszewski, L. M. & Bohmann, D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78, 787–798 (1994).
Dart, A. E. et al. PAK4 promotes kinase-independent stabilization of RhoU to modulate cell adhesion. J. Cell. Biol. 211, 863–879 (2015).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Schaller, T. et al. Effects of inner nuclear membrane proteins SUN1/UNC-84A and SUN2/UNC-84B on the EARLY steps of HIV-1 infection. J. Virol. 91, e00463-17 (2017).
Matheson, N. J., Peden, A. A. & Lehner, P. J. Antibody-free magnetic cell sorting of genetically modified primary human CD4+T cells by one-step streptavidin affinity purification. PLoS ONE 9, e111437 (2014).
Pertel, T. et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365 (2011).
Acknowledgements
We thank D. Pollpeter, M. Dicks, S. Papaioannou, C. Wells and S. Wolinsky for the generous provision of reagents and helpful discussions. The work was supported by the UK Medical Research Council (grant no. G1000196), the Wellcome Trust (grant no. 106223/Z/14/Z), and the Department of Health via a National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s and St. Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust. J.M.J.-G. is a Long-Term Fellow of the European Molecular Biology Organization (EMBO) (ALTF 663-2016).
Author information
Authors and Affiliations
Contributions
J.M.J.-G., L.A., and M.H.M. conceived the siRNA screen. J.M.J.-G. and M.H.M. designed the study and wrote the manuscript with input from all co-authors. J.M.J.-G. carried out the experiments and analysed the data. L.A. and G.B. contributed to the execution of experiments and provided reagents. M.H.M. supervised all aspects of the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–24 and Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Jimenez-Guardeño, J.M., Apolonia, L., Betancor, G. et al. Immunoproteasome activation enables human TRIM5α restriction of HIV-1. Nat Microbiol 4, 933–940 (2019). https://doi.org/10.1038/s41564-019-0402-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0402-0
This article is cited by
-
Autophagy-enhancing ATG16L1 polymorphism is associated with improved clinical outcome and T-cell immunity in chronic HIV-1 infection
Nature Communications (2024)
-
TRIM5α restricts poxviruses and is antagonized by CypA and the viral protein C6
Nature (2023)
-
TRIM5α recruits HDAC1 to p50 and Sp1 and promotes H3K9 deacetylation at the HIV-1 LTR
Nature Communications (2023)
-
HIV-1 capsid variability: viral exploitation and evasion of capsid-binding molecules
Retrovirology (2021)
-
MX2-mediated innate immunity against HIV-1 is regulated by serine phosphorylation
Nature Microbiology (2021)