Abstract
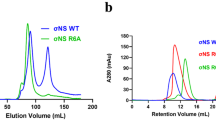
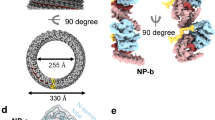
Bluetongue virus (BTV) non-structural protein 1 (NS1) regulates viral protein synthesis and exists as tubular and non-tubular forms in infected cells, but how tubules assemble and how protein synthesis is regulated are unknown. Here, we report near-atomic resolution structures of two NS1 tubular forms determined by cryo-electron microscopy. The two tubular forms are different helical assemblies of the same NS1 monomer, consisting of an amino-terminal foot, a head and body domains connected to an extended carboxy-terminal arm, which wraps atop the head domain of another NS1 subunit through hydrophobic interactions. Deletion of the C terminus prevents tubule formation but not viral replication, suggesting an active non-tubular form. Two zinc-finger-like motifs are present in each NS1 monomer, and tubules are disrupted by divalent cation chelation and restored by cation addition, including Zn2+, suggesting a regulatory role of divalent cations in tubule formation. In vitro luciferase assays show that the NS1 non-tubular form upregulates BTV mRNA translation, whereas zinc-finger disruption decreases viral mRNA translation, tubule formation and virus replication, confirming a functional role for the zinc-fingers. Thus, the non-tubular form of NS1 is sufficient for viral protein synthesis and infectious virus replication, and the regulatory mechanism involved operates through divalent cation-dependent conversion between the non-tubular and tubular forms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Walsh, D. & Mohr, I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 9, 860–875 (2011).
Piron, M., Vende, P., Cohen, J. & Poncet, D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17, 5811–5821 (1998).
Piron, M., Delaunay, T., Grosclaude, J. & Poncet, D. Identification of the RNA-binding, dimerization, and eIF4GI-binding domains of rotavirus nonstructural protein NSP3. J. Virol. 73, 5411–5421 (1999).
Boyce, M., Celma, C. P. & Roy, P. Bluetongue virus non-structural protein 1 is a positive regulator of viral protein synthesis. Virol. J. 9, 178 (2012).
Vitour, D., Lindenbaum, P., Vende, P., Becker, M. M. & Poncet, D. RoXaN, a novel cellular protein containing TPR, LD, and zinc finger motifs, forms a ternary complex with eukaryotic initiation factor 4G and rotavirus NSP3. J. Virol. 78, 3851–3862 (2004).
Harb, M. et al. Nuclear localization of cytoplasmic poly(A)-binding protein upon rotavirus infection involves the interaction of NSP3 with eIF4G and RoXaN. J. Virol. 82, 11283–11293 (2008).
Deo, R. C., Groft, C. M., Rajashankar, K. R. & Burley, S. K. Recognition of the rotavirus mRNA 3′ consensus by an asymmetric NSP3 homodimer. Cell 108, 71–81 (2002).
Groft, C. M. & Burley, S. K. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol. Cell 9, 1273–1283 (2002).
Hewat, E. A., Booth, T. F., Wade, R. H. & Roy, P. 3-D reconstruction of bluetongue virus tubules using cryoelectron microscopy. J. Struct. Biol. 108, 35–48 (1992).
Urakawa, T. & Roy, P. Bluetongue virus tubules made in insect cells by recombinant baculoviruses: expression of the NS1 gene of bluetongue virus serotype 10. J. Virol. 62, 3919–3927 (1988).
Odom, T. W., Huang, J. L. & Lieber, C. M. Single-walled carbon nanotubes: from fundamental studies to new device concepts. Ann. N. Y. Acad. Sci. 960, 203–215 (2002).
Monastyrskaya, K., Gould, E. A. & Roy, P. Characterization and modification of the carboxy-terminal sequences of bluetongue virus type 10 NS1 protein in relation to tubule formation and location of an antigenic epitope in the vicinity of the carboxy terminus of the protein. J. Virol. 69, 2831–2841 (1995).
Boyce, M., Celma, C. C. & Roy, P. Development of reverse genetics systems for bluetongue virus: recovery of infectious virus from synthetic RNA transcripts. J. Virol. 82, 8339–8348 (2008).
Monastyrskaya, K., Booth, T., Nel, L. & Roy, P. Mutation of either of 2 cysteine residues or deletion of the amino or carboxy-terminus of nonstructural protein Ns1 of bluetongue virus abrogates virus-specified tubule formation in insect cells. J. Virol. 68, 2169–2178 (1994).
Mikhailov, M., Monastyrskaya, K., Bakker, T. & Roy, P. A new form of particulate single and multiple immunogen delivery system based on recombinant bluetongue virus-derived tubules. Virology 217, 323–331 (1996).
Ghosh, M. K., Borca, M. V. & Roy, P. Virus-derived tubular structure displaying foreign sequences on the surface elicit CD4+ Th cell and protective humoral responses. Virology 302, 383–392 (2002).
Murphy, A. & Roy, P. Manipulation of the bluetongue virus tubules for immunogen delivery. Future Microbiol. 3, 351–359 (2008).
Owens, R. J., Limn, C. & Roy, P. Role of an arbovirus nonstructural protein in cellular pathogenesis and virus release. J. Virol. 78, 6649–6656 (2004).
Mortola, E., Noad, R. & Roy, P. Bluetongue virus outer capsid proteins are sufficient to trigger apoptosis in mammalian cells. J. Virol. 78, 2875–2883 (2004).
Hall, T. M. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 15, 367–373 (2005).
Laity, J. H., Lee, B. M. & Wright, P. E. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46 (2001).
Attar, N. et al. The histone H3–H4 tetramer is a copper reductase enzyme. Preprint at https://www.biorxiv.org/content/early/2018/06/19/350652 (2018).
Yu, X., Jiang, J., Sun, J. & Zhou, Z. H. A putative ATPase mediates RNA transcription and capping in a dsRNA virus. eLife 4, e07901 (2015).
Carragher, B. et al. Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 132, 33–45 (2000).
Ge, P. & Zhou, Z. H. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc. Natl Acad. Sci. USA 108, 9637–9642 (2011).
Zhang, X., Jin, L., Fang, Q., Hui, W. H. & Zhou, Z. H. 3.3 Å cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141, 472–482 (2010).
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semi-automated software for high resolution single particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Diaz, R., Rice, W. J. & Stokes, D. L. Fourier–Bessel reconstruction of helical assemblies. Methods Enzymol. 482, 131–165 (2010).
Egelman, E. H. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J. Struct. Biol. 157, 83–94 (2007).
Ge, P. et al. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat. Struct. Mol. Biol. 22, 377–382 (2015).
Clemens, D. L., Ge, P., Lee, B. Y., Horwitz, M. A. & Zhou, Z. H. Atomic structure of T6SS reveals interlaced array essential to function. Cell 160, 940–951 (2015).
Bartesaghi, A. et al. 2.2 Å resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science 348, 1147–1151 (2015).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
We thank M. Turmaine for his advice and support for imaging at the UCL EM facility and C. Celma (LSHTM) for advising in the BTV reverse genetics method. This project is supported partly by grants from the US NIH (AI094386 to Z.H.Z.) and The Wellcome Trust, UK (100218, Investigator Award to P.R.). We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and grants from the NIH (1S10OD018111 and 1U24 GM116792) and the National Science Foundation (DBI-1338135 and DMR-1548924). This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation grant number ACI-1548562 (Comet cluster at the San Diego Supercomputing Center through allocation MCB140140).
Author information
Authors and Affiliations
Contributions
Z.H.Z., P.R. and P.G. designed the experiments. M.B. purified the wild-type NS1 tubules. X.Z. recorded some of the cryoEM data. P.G. recorded the cryoEM data and determined the structure. M.L. and J.J. built the atomic models. A.K. expressed proteins, performed the mutagenesis and biochemical experiments, reverse genetics, virology and fluorescence microscopy analyses. M.L., Z.H.Z., P.R., P.G. and A.K. interpreted the data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–4 and Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Kerviel, A., Ge, P., Lai, M. et al. Atomic structure of the translation regulatory protein NS1 of bluetongue virus. Nat Microbiol 4, 837–845 (2019). https://doi.org/10.1038/s41564-019-0369-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0369-x
This article is cited by
-
Role of environmental specificity in CASP results
BMC Bioinformatics (2023)