Abstract
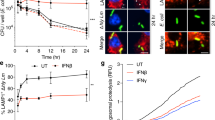
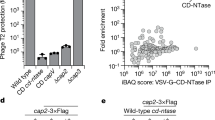
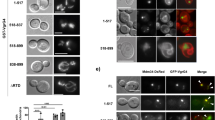
The innate immune system is crucial for eventual control of infections, but may also contribute to pathology. Listeria monocytogenes is an intracellular Gram-positive bacteria and a major cause of food-borne disease. However, important knowledge on the interactions between L. monocytogenes and the immune system is still missing. Here, we report that Listeria DNA is sorted into extracellular vesicles (EVs) in infected cells and delivered to bystander cells to stimulate the cyclic guanosine monophosphate–adenosine monophosphate synthase (cGAS)–stimulator of interferon genes (STING) pathway. This was also observed during infections with Francisella tularensis and Legionella pneumophila. We identify the multivesicular body protein MVB12b as a target for TANK-binding kinase 1 phosphorylation, which is essential for the sorting of DNA into EVs and stimulation of bystander cells. EVs from Listeria-infected cells inhibited T-cell proliferation, and primed T cells for apoptosis. Collectively, we describe a pathway for EV-mediated delivery of foreign DNA to bystander cells, and suggest that intracellular bacteria exploit this pathway to impair antibacterial defence.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The full next-generation sequencing dataset is available at the European Nucleotide Archive with the identifier ‘ena-STUDY-AARHUS UNIVERSITY 12-12-2018-17:12:31:528-124’, under accession number PRJEB30324. The full mass spectrometry dataset is available at https://www.ebi.ac.uk/pride/archive/projects/PXD012135. Original immunoblots are shown in Supplementary Fig. 7.
References
Barbuddhe, S. B. & Chakraborty, T. Listeria as an enteroinvasive gastrointestinal pathogen. Curr. Top. Microbiol. Immunol. 337, 173–195 (2009).
Cossart, P. et al. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect. Immun. 57, 3629–3636 (1989).
Rothe, J. et al. Mice lacking the tumour necrosis factor receptor 1 are resistant to TNF-mediated toxicity but highly susceptible to infection by Listeria monocytogenes. Nature 364, 798–802 (1993).
Edelson, B. T. & Unanue, E. R. MyD88-dependent but Toll-like receptor 2-independent innate immunity to Listeria: no role for either in macrophage listericidal activity. J. Immunol. 169, 3869–3875 (2002).
Ladel, C. H., Flesch, I. E., Arnoldi, J. & Kaufmann, S. H. Studies with MHC-deficient knock-out mice reveal impact of both MHC I- and MHC II-dependent T cell responses on Listeria monocytogenes infection. J. Immunol. 153, 3116–3122 (1994).
Stockinger, S. et al. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J. Immunol. 169, 6522–6529 (2002).
Auerbuch, V., Brockstedt, D. G., Meyer-Morse, N., O’Riordan, M. & Portnoy, D. A. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 200, 527–533 (2004).
Carrero, J. A., Calderon, B. & Unanue, E. R. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J. Exp. Med. 200, 535–540 (2004).
O’Connell, R. M. et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J. Exp. Med. 200, 437–445 (2004).
Luecke, S. & Paludan, S. R. Molecular requirements for sensing of intracellular microbial nucleic acids by the innate immune system. Cytokine 98, 4–14 (2016).
Kawai, T. & Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 (2011).
Kawai, T. et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 (2005).
Seth, R. B., Sun, L., Ea, C. K. & Chen, Z. J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682 (2005).
Ishikawa, H., Ma, Z. & Barber, G. N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Berg, R. K. et al. T cells detect intracellular DNA but fail to induce type I IFN responses: implications for restriction of HIV replication. PLoS ONE 9, e84513 (2014).
Larkin, B. et al. Cutting edge: activation of STING in T cells induces type I IFN responses and cell death. J. Immunol. 199, 397–402 (2017).
Cerboni, S. et al. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J. Exp. Med. 214, 1769–1785 (2017).
Gulen, M. F. et al. Signalling strength determines proapoptotic functions of STING. Nat. Commun. 8, 427 (2017).
Hansen, K. et al. Listeria monocytogenes induces IFNβ expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 33, 1654–1666 (2014).
Woodward, J. J., Iavarone, A. T. & Portnoy, D. A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705 (2010).
Abdullah, Z. et al. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 31, 4153–4164 (2012).
Czuczman, M. A. et al. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature 509, 230–234 (2014).
Schorey, J. S., Cheng, Y., Singh, P. P. & Smith, V. L. Exosomes and other extracellular vesicles in host–pathogen interactions. EMBO Rep. 16, 24–43 (2015).
Trajkovic, K. et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 (2008).
Kosaka, N. et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452 (2010).
Liu, S. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015).
Jeppesen, D. K. et al. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J. Extracell. Vesicles 3, 25011 (2014).
Burdette, D. L. et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011).
Ong, S. E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002).
Christ, L., Raiborg, C., Wenzel, E. M., Campsteijn, C. & Stenmark, H. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci. 42, 42–56 (2017).
Ablasser, A. et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534 (2013).
Bridgeman, A. et al. Viruses transfer the antiviral second messenger cGAMP between cells. Science 349, 1228–1232 (2015).
Gentili, M. et al. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science 349, 1232–1236 (2015).
Robbins, P. D. & Morelli, A. E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 14, 195–208 (2014).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Dreux, M. et al. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe 12, 558–570 (2012).
Yang, X. et al. Hepatitis B virus-encoded microRNA controls viral replication. J. Virol. 91, e01919–16 (2017).
Pegtel, D. M. et al. Functional delivery of viral miRNAs via exosomes. Proc. Natl Acad. Sci. USA 107, 6328–6333 (2010).
Henry, T. et al. Type I IFN signaling constrains IL-17A/F secretion by γδ T cells during bacterial infections. J. Immunol. 184, 3755–3767 (2010).
Gaidt, M. M. et al. The DNA inflammasome in human myeloid cells is initiated by a STING–cell death program upstream of NLRP3. Cell 171, 1110–1124 (2017).
Paludan, S. R. & Bowie, A. G. Immune sensing of DNA. Immunity 38, 870–880 (2013).
Fernandes-Alnemri, T. et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11, 385–393 (2010).
Kim, S. et al. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 40, 1545–1551 (2010).
Stockinger, S. et al. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS Pathog. 5, e1000355 (2009).
Holm, C. K. et al. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat. Commun. 7, 10680 (2016).
Ogunjimi, B. et al. Inborn errors in RNA polymerase III underlie severe Varicella zoster virus infections. J. Clin. Invest. 127, 3543–3556 (2017).
Skoble, J., Portnoy, D. A. & Welch, M. D. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J. Cell Biol. 150, 527–538 (2000).
Weiss, D. S. et al. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl Acad. Sci. USA 104, 6037–6042 (2007).
Horan, K. A. et al. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J. Immunol. 190, 2311–2319 (2013).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Thery, C., Amigorena, S., Raposo, G. & Clayton, A. in Current Protocols in Cell Biology 3.22.1–3.22.29 (Wiley Online Library, 2006).
Chevillet, J. R. et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl Acad. Sci. USA 111, 14888–14893 (2014).
Acknowledgements
The technical assistance of K. Stadel Petersen, the next-generation sequencing facility at the Department of Molecular Medicine, Aarhus University Hospital, and the FACS Core facility, Aarhus University is greatly appreciated. This work was funded by The Danish Medical Research Council (12-124330 to S.R.P.), Novo Nordisk Foundation (NNF18OC0030274 to S.R.P.), Lundbeck Foundation (R198-2015-171 to S.R.P.), European Research Council (786602 to S.R.P.), EU FP7 MOBILEX programme (DFF – 5053-00011 to R.N.), BMBF (JPI-AMR - FKZ 01Kl1702) and DFG (SFB/TR-84 TP C01) (both to B.S.), and Austrian Science Fund through grant P 25186-B22 (to T.D.).
Author information
Authors and Affiliations
Contributions
R.N. and S.R.P. conceived the idea and designed the experiments. R.N., R.T., F.M., T.P., A.K., B.Z, S.A., A.M., W.B., A.A., T.K. and M.K.T. performed the experiments. F.M. designed, performed and analysed the phosphoproteomics experiment. E.F. analysed the next-generation sequencing data. C.K.H., B.S., K.A.H., T.H., K.V.G., T.D. and S.R.P. supervised the experiments. R.N. and S.R.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–7 and Supplementary Table 1.
Supplementary Table 2
Full dataset for phosphoproteome analysis of dsDNA-stimulated cells.
Rights and permissions
About this article
Cite this article
Nandakumar, R., Tschismarov, R., Meissner, F. et al. Intracellular bacteria engage a STING–TBK1–MVB12b pathway to enable paracrine cGAS–STING signalling. Nat Microbiol 4, 701–713 (2019). https://doi.org/10.1038/s41564-019-0367-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0367-z
This article is cited by
-
S-nitrosothiol homeostasis maintained by ADH5 facilitates STING-dependent host defense against pathogens
Nature Communications (2024)
-
STING dependent BAX-IRF3 signaling results in apoptosis during late-stage Coxiella burnetii infection
Cell Death & Disease (2024)
-
Eosinophils promote CD8+ T cell memory generation to potentiate anti-bacterial immunity
Signal Transduction and Targeted Therapy (2024)
-
SAM68 directs STING signaling to apoptosis in macrophages
Communications Biology (2024)
-
Exosome, the glass slipper for Cinderella of cancer—bladder cancer?
Journal of Nanobiotechnology (2023)