Abstract
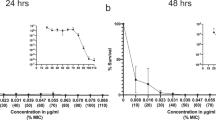
When choosing antibiotics to treat bacterial infections, it is assumed that the susceptibility of the target bacteria to an antibiotic is reflected by laboratory estimates of the minimum inhibitory concentration (MIC) needed to prevent bacterial growth. A caveat of using MIC data for this purpose is heteroresistance, the presence of a resistant subpopulation in a main population of susceptible cells. We investigated the prevalence and mechanisms of heteroresistance in 41 clinical isolates of the pathogens Escherichia coli, Salmonella enterica, Klebsiella pneumoniae and Acinetobacter baumannii against 28 different antibiotics. For the 766 bacteria–antibiotic combinations tested, as much as 27.4% of the total was heteroresistant. Genetic analysis demonstrated that a majority of heteroresistance cases were unstable, with an increased resistance of the subpopulations resulting from spontaneous tandem amplifications, typically including known resistance genes. Using mathematical modelling, we show how heteroresistance in the parameter range estimated in this study can result in the failure of antibiotic treatment of infections with bacteria that are classified as antibiotic susceptible. The high prevalence of heteroresistance with the potential for treatment failure highlights the limitations of MIC as the sole criterion for susceptibility determinations. These results call for the development of facile and rapid protocols to identify heteroresistance in pathogens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Chromosomes and plasmids are deposited at NCBI under the following accession numbers: DA33098 (CP029569–CP029573), DA33133 (CP029574 and CP029575), DA33135 (CP029576–CP029578), DA33137 (CP029579–CP029581), DA33140 (CP029582–CP029586), DA33141 (CP029587–CP029589), DA33144 (CP029590–CP029592), DA33145 (CP029597–CP029599), DA33382 (CP030106–CP030109), DA34821 (CP029567), DA34827 (CP029593 and CP029594), DA34833 (CP029595 and CP029596) and DA34837 (CP029568). Additional data supporting the findings of this study, such as raw data and bacterial strains, are available upon request.
References
The Bacterial Challenge – Time to React ECDC/ EMEA Joint Technical Report (ECDC/EMEA Joint Working Group, 2009).
Antibiotic Resistance Threats in the United States (Centers for Disease Control and Prevention, Office of Infectious Diseases, 2013).
Frimodt-Møller, N. How predictive is PK/PD for antibacterial agents? Int. J. Antimicrob. Agents 19, 333–339 (2002).
Asín-Prieto, E., Rodríguez-Gascón, A. & Isla, A. Applications of the pharmacokinetic/pharmacodynamic (PK/PD) analysis of antimicrobial agents. J. Infect. Chemother. 21, 319–329 (2015).
Nielsen, E. I. & Friberg, L. E. Pharmacokinetic-pharmacodynamic modeling of antibacterial drugs. Pharmacol. Rev. 65, 1053–1090 (2013).
El-Halfawy, O. M. & Valvano, M. A. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207 (2015).
Alexander, H. E. & Leidy, G. Mode of action of streptomycin on type b Haemophilus influenzae: I. Origin of resistant organisms. J. Exp. Med. 85, 329–338 (1947).
Band, V. I. et al. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat. Microbiol. 1, 16053 (2016).
Higgins, P. G., Schneiders, T., Hamprecht, A. & Seifert, H. In vivo selection of a missense mutation in adeR and conversion of the novel blaOXA-164 gene into blaOXA-58 in carbapenem-resistant Acinetobacter baumannii isolates from a hospitalized patient. Antimicrob. Agents Chemother. 54, 5021–5027 (2010).
Hernan, R. C. et al. Selection of colistin-resistant Acinetobacter baumannii isolates in postneurosurgical meningitis in an intensive care unit with high presence of heteroresistance to colistin. Diagn. Microbiol. Infect. Dis. 65, 188–191 (2009).
Tan, C.-H., Li, J. & Nation, R. L. Activity of colistin against heteroresistant Acinetobacter baumannii and emergence of resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob. Agents Chemother. 51, 3413–3415 (2007).
Band, V. I. et al. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 9, e02448–17 (2018).
Moore, M. R., Perdreau-Remington, F. & Chambers, H. F. Vancomycin treatment failure associated with heterogeneous vancomycin-intermediate Staphylococcus aureus in a patient with endocarditis and in the rabbit model of endocarditis. Antimicrob. Agents Chemother. 47, 1262–1266 (2003).
Yau, W. et al. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the Western Pacific region in the SENTRY antimicrobial surveillance programme. J. Infect. 58, 138–144 (2009).
Meletis, G., Tzampaz, E., Sianou, E., Tzavaras, I. & Sofianou, D. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 66, 946–947 (2011).
Sun, J. D. et al. Impact of carbapenem heteroresistance among clinical isolates of invasive Escherichia coli in Chongqing, southwestern China. Clin. Microbiol. Infect. 21, 469.e1–469.e10 (2015).
Mei, S., Gao, Y., Zhu, C., Dong, C. & Chen, Y. Research of the heteroresistance of Pseudomonas aeruginosa to imipenem. Int. J. Clin. Exp. Med. 8, 6129–6132 (2015).
van Hal, S. J. et al. Performance of various testing methodologies for detection of heteroresistant vancomycin-intermediate Staphylococcus aureus in bloodstream isolates. J. Clin. Microbiol. 49, 1489–1494 (2011).
Lo-Ten-Foe, J. R., de Smet, A. M. G. A., Diederen, B. M. W., Kluytmans, J. A. J. W. & van Keulen, P. H. J. Comparative evaluation of the VITEK 2, disk diffusion, etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3726–3730 (2007).
Lee, H.-Y. et al. Imipenem heteroresistance induced by imipenem in multidrug-resistant Acinetobacter baumannii: mechanism and clinical implications. Int. J. Antimicrob. Agents 37, 302–308 (2011).
Hjort, K., Nicoloff, H. & Andersson, D. I. Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol. Microbiol. 102, 274–289 (2016).
Halaby, T. et al. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob. Agents Chemother. 60, 6837–6843 (2016).
Jayol, A., Nordmann, P., Brink, A. & Poirel, L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 59, 2780–2784 (2015).
Schechter, L. M. et al. Extensive gene amplification as a mechanism for piperacillin-tazobactam resistance in Escherichia coli. mBio 9, e00583-18 (2018).
Williams, A. B. Spontaneous mutation rates come into focus in Escherichia coli. DNA Repair 24, 73–79 (2014).
Lee, J.-Y., Choi, M.-J., Choi, H. J. & Ko, K. S. Preservation of acquired colistin resistance in Gram-negative bacteria. Antimicrob. Agents Chemother. 60, 609–612 (2015).
Barin, J., Martins, A. F., Heineck, B. L., Barth, A. L. & Zavascki, A. P. Hetero- and adaptive resistance to polymyxin B in OXA-23-producing carbapenem-resistant Acinetobacter baumannii isolates. Ann. Clin. Microbiol. Antimicrob. 12, 15 (2013).
Poudyal, A. et al. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 62, 1311–1318 (2008).
Cai, Y., Chai, D., Wang, R., Liang, B. & Bai, N. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67, 1607–1615 (2012).
Reams, A. B. & Roth, J. R. Mechanisms of gene duplication and amplification. Cold Spring Harb. Perspect. Biol. 7, a016592 (2015).
Ramiro, R. S., Costa, H. & Gordo, I. Macrophage adaptation leads to parallel evolution of genetically diverse Escherichia coli small-colony variants with increased fitness in vivo and antibiotic collateral sensitivity. Evol. Appl. 9, 994–1004 (2016).
Paradise, M. R., Cook, G., Poole, R. K. & Rather, P. N. Mutations in aarE, the ubiA homolog of Providencia stuartii, result in high-level aminoglycoside resistance and reduced expression of the chromosomal aminoglycoside 2′-N-acetyltransferase. Antimicrob. Agents Chemother. 42, 959–962 (1998).
Gallagher, L. A., Lee, S. A. & Manoil, C. Importance of core genome functions for an extreme antibiotic resistance trait. mBio 8, e01655-17 (2017).
Shan, Y., Lazinski, D., Rowe, S., Camilli, A. & Lewis, K. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio 6, e00078-15 (2015).
Tian, Z.-X., Yi, X.-X., Cho, A., O’Gara, F. & Wang, Y.-P. CpxR activates MexAB-OprM efflux pump expression and enhances antibiotic resistance in both laboratory and clinical nalB-type isolates of Pseudomonas aeruginosa. PLoS Pathog. 12, e1005932 (2016).
Weatherspoon-Griffin, N., Yang, D., Kong, W., Hua, Z. & Shi, Y. The CpxR/CpxA two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide. J. Biol. Chem. 289, 32571–32582 (2014).
Shalel-Levanon, S., San, K.-Y. & Bennett, G. N. Effect of oxygen, and ArcA and FNR regulators on the expression of genes related to the electron transfer chain and the TCA cycle in Escherichia coli. Metab. Eng. 7, 364–374 (2005).
Coyne, S., Courvalin, P. & Périchon, B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55, 947–953 (2011).
El Meouche, I., Siu, Y. & Dunlop, M. J. Stochastic expression of a multiple antibiotic resistance activator confers transient resistance in single cells. Sci. Rep. 6, 19538 (2016).
Sanchez-Romero, M. A. & Casadesus, J. Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc. Natl Acad. Sci. USA 111, 355–360 (2014).
Allen, R. C., Engelstädter, J., Bonhoeffer, S., McDonald, B. A. & Hall, A. R. Reversing resistance: different routes and common themes across pathogens. P. Roy. Soc. B-Biol. Sci. 284, 20171619 (2017).
Anderson, P. & Roth, J. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc. Natl Acad. Sci. USA 78, 3113–3117 (1981).
Sandegren, L. & Andersson, D. I. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7, 578–588 (2009).
Reams, A. B., Kofoid, E., Savageau, M. & Roth, J. R. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics 184, 1077–1094 (2010).
Adler, M., Anjum, M., Berg, O. G., Andersson, D. I. & Sandegren, L. High fitness costs and instability of gene duplications reduce rates of evolution of new genes by duplication-divergence mechanisms. Mol. Biol. Evol. 31, 1526–1535 (2014).
Sun, S., Berg, O. G., Roth, J. R. & Andersson, D. I. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics 182, 1183–1195 (2009).
Regoes, R. R. et al. Pharmacodynamic functions: a multiparameter approach to the design of antibiotic treatment regimens. Antimicrob. Agents Chemother. 48, 3670–3676 (2004).
Kussell, E., Kishony, R., Balaban, N. Q. & Leibler, S. Bacterial persistence: a model of survival in changing environments. Genetics 169, 1807–1814 (2005).
Johnson, P. J. T. & Levin, B. R. Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genet. 9, e1003123 (2013).
Levin, B. R., Concepción-Acevedo, J. & Udekwu, K. I. Persistence: a copacetic and parsimonious hypothesis for the existence of non-inherited resistance to antibiotics. Curr. Opin. Microbiol. 21, 18–21 (2014).
Anderson, S. E., Sherman, E. X., Weiss, D. S. & Rather, P. N. Aminoglycoside heteroresistance in Acinetobacter baumannii AB5075. mSphere 3, e00271-18 (2018).
Levin, B. R., Baquero, F., Ankomah, P. & McCall, I. C. Phagocytes, antibiotics, and self-limiting bacterial infections. Trends Microbiol. 25, 878–892 (2017).
Wootton, M. et al. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47, 399–403 (2001).
Sutton, S. Measurement of microbial cells by optical density. J. Valid. Technol. 17, 46–49 (2011).
Udekwu, K. I., Parrish, N., Ankomah, P., Baquero, F. & Levin, B. R. Functional relationship between bacterial cell density and the efficacy of antibiotics. J. Antimicrob. Chemother. 63, 745–757 (2009).
Thulin, M. BAT v.2.0 (Uppsala University, 2018); http://www.mansthulin.se/bat/
Nicoloff, H. & Andersson, D. I. Indirect resistance to several classes of antibiotics in cocultures with resistant bacteria expressing antibiotic-modifying or -degrading enzymes. J. Antimicrob. Chemother. 71, 100–110 (2016).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644 (2012).
Acknowledgements
We thank F.F. Cuenca, P.G. Higgins and C.-H. Chiu for the A. baumannii isolates, L. Sandegren for the E. coli isolates, Å. Melhus and B. Lytsy for some K. pneumoniae isolates and C. Järnberg at The Public Health Agency, Sweden for the S. Typhimurium isolates. We also would like to thank O. Warsi and U. Lustig for helping with the MiSeq sequencer and DNA libraries preparations. This work was supported by grants from the Swedish Research Council, no. 2017-01527 (DIA), and the US National Institutes of General Medical Sciences, no. GM091875 (BRL).
Author information
Authors and Affiliations
Contributions
H.N., K.H. and D.I.A. designed the study. H.N. and K.H. performed the experiments and B.R.L. the mathematical modelling. H.N., K.H., B.R.L. and D.I.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–12, Supplementary Tables 1–5, Supplementary Methods and Supplementary References.
Rights and permissions
About this article
Cite this article
Nicoloff, H., Hjort, K., Levin, B.R. et al. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol 4, 504–514 (2019). https://doi.org/10.1038/s41564-018-0342-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0342-0
This article is cited by
-
Bacteria can compensate the fitness costs of amplified resistance genes via a bypass mechanism
Nature Communications (2024)
-
Cephalosporin resistance, tolerance, and approaches to improve their activities
The Journal of Antibiotics (2024)
-
Plasmid-mediated phenotypic noise leads to transient antibiotic resistance in bacteria
Nature Communications (2024)
-
Duplicated antibiotic resistance genes reveal ongoing selection and horizontal gene transfer in bacteria
Nature Communications (2024)
-
Emerging tools for uncovering genetic and transcriptomic heterogeneities in bacteria
Biophysical Reviews (2024)