Abstract
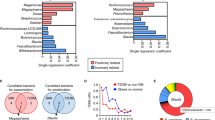
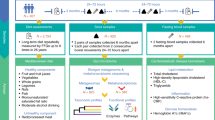
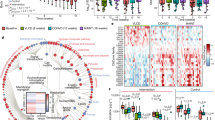
Humans with metabolic and inflammatory diseases frequently harbour lower levels of butyrate-producing bacteria in their gut. However, it is not known whether variation in the levels of these organisms is causally linked with disease development and whether diet modifies the impact of these bacteria on health. Here we show that a prominent gut-associated butyrate-producing bacterial genus (Roseburia) is inversely correlated with atherosclerotic lesion development in a genetically diverse mouse population. We use germ-free apolipoprotein E-deficient mice colonized with synthetic microbial communities that differ in their capacity to generate butyrate to demonstrate that Roseburia intestinalis interacts with dietary plant polysaccharides to: impact gene expression in the intestine, directing metabolism away from glycolysis and toward fatty acid utilization; lower systemic inflammation; and ameliorate atherosclerosis. Furthermore, intestinal administration of butyrate reduces endotoxaemia and atherosclerosis development. Together, our results illustrate how modifiable diet-by-microbiota interactions impact cardiovascular disease, and suggest that interventions aimed at increasing the representation of butyrate-producing bacteria may provide protection against atherosclerosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability:
The SRA accession ID for COPRO-Seq is SRP158926. The accession number for RNA sequencing data is GEO: GSE119141. The SRA accession ID for the Ath-HMDP microbiome data is SRP142550. Additional data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L. & Gordon, J. I. Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011).
Hooper, L. V., Midtvedt, T. & Gordon, J. I. How host–microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307 (2002).
Xu, J. et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 5, e156 (2007).
Martens, E. C., Koropatkin, N. M., Smith, T. J. & Gordon, J. I. Complex glycan catabolism by the human gut microbiota: the bacteroidetes sus-like paradigm. J. Biol. Chem. 284, 24673–24677 (2009).
Koropatkin, N. M., Cameron, E. A. & Martens, E. C. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10, 323–335 (2012).
Martens, E. C., Kelly, A. G., Tauzin, A. S. & Brumer, H. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J. Mol. Biol. 426, 3851–3865 (2014).
Schnorr, S. et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5, 3654 (2014).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Ritzhaupt, A., Ellis, A., Hosie, K. B. & Shirazi-Beechey, S. P. The characterization of butyrate transport across pig and human colonic luminal membrane. J. Physiol. 507, 819–830 (1998).
Candido, E. P. M., Reeves, R. & Davie, J. R. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell 14, 105–113 (1978).
Segain, J. P. et al. Butyrate inhibits inflammatory responses through NFκB inhibition: implications for Crohn’s disease. Gut 47, 397–403 (2000).
Kasahara, K. et al. Commensal bacteria at the crossroad between cholesterol homeostasis and chronic inflammation in atherosclerosis. J. Lipid Res. 58, 519–528 (2017).
Mitchell, J. A., Ryffel, B., Quesniaux, V. F. J., Cartwright, N. & Paul-Clark, M. Role of pattern-recognition receptors in cardiovascular health and disease. Biochem. Soc. Trans. 35, 1449–1452 (2007).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–65 (2011).
Tang, W. H. W. et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584 (2013).
Karlsson, F. H. et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 3, 1245 (2012).
Louis, P., Young, P., Holtrop, G. & Flint, H. J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 12, 304–314 (2010).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
Karlsson, F. H. et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013).
Walker, A. W. et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 5, 220–230 (2011).
Duncan, S. H. et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 73, 1073–1078 (2007).
Bennett, B. J. et al. Genetic architecture of atherosclerosis in mice: a systems genetics analysis of common inbred strains. PLoS Genet. 11, e1005711 (2015).
Duncan, S. H., Barcenilla, A., Stewart, C. S., Pryde, S. E. & Flint, H. J. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 68, 5186–5190 (2002).
Guilloteau, P. et al. From the gut to the peripheral tissues: the multiple effects of butyrate. Nutr. Res. Rev. 23, 366–384 (2010).
Cybulsky, M. I. et al. A major role for VCAM-1, but not ICAM-1, in early atherosclerosis. J. Clin. Invest. 107, 1255–1262 (2001).
Boesten, L. S. M. et al. Tumor necrosis factor-α promotes atherosclerotic lesion progression in APOE*3-leiden transgenic mice. Cardiovasc. Res. 66, 179–185 (2005).
Nightingale, K. P. et al. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J. Biol. Chem. 282, 4408–4416 (2007).
Fan, J., Krautkramer, K. A., Feldman, J. L. & Denu, J. M. Metabolic regulation of histone post-translational modifications. ACS Chem. Biol. 10, 95–108 (2015).
Krautkramer, K. A., Reiter, L., Denu, J. M. & Dowell, J. A. Quantification of SAHA-dependent changes in histone modifications using data-independent acquisition mass spectrometry. J. Proteome Res. 14, 3252–3262 (2015).
Donohoe, D. R. et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13, 517–526 (2011).
Peng, L., Li, Z.-R., Green, R. S., Holzman, I. R. & Lin, J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 139, 1619–1625 (2009).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Ohira, H. et al. Butyrate attenuates inflammation and lipolysis generated by the interaction of adipocytes and macrophages. J. Atheroscler. Thromb. 20, 425–442 (2013).
Li, H. et al. Sodium butyrate stimulates expression of fibroblast growth factor 21 in liver by inhibition of histone deacetylase 3. Diabetes 61, 797–806 (2012).
Youm, Y. H. et al. The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat. Med. 21, 263–269 (2015).
Balsinde, J., Balboa, M. A. & Dennis, E. A. Inflammatory activation of arachidonic acid signaling in murine P388D1 macrophages via sphingomyelin synthesis. J. Biol. Chem. 272, 20373–20377 (1997).
Zhao, L. et al. Selective interleukin-12 synthesis defect in 12/15-lipoxygenase-deficient macrophages associated with reduced atherosclerosis in a mouse model of familial hypercholesterolemia. J. Biol. Chem. 277, 35350–35356 (2002).
Cyrus, T. et al. Disruption of the 12/15-lipoxygenase gene diminishes atherosclerosis in apo E-deficient mice. J. Clin. Invest. 103, 1597–1604 (1999).
Threapleton, D. E. et al. Dietary fibre intake and risk of cardiovascular disease: systematic review and meta-analysis. BMJ 347, f6879 (2013).
Marques, F. Z. et al. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation 135, 964–977 (2017).
Rivière, A., Selak, M., Lantin, D., Leroy, F. & De Vuyst, L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 7, 979 (2016).
Ait-Oufella, H. et al. Natural regulatory T cells control the development of atherosclerosis in mice. Nat. Med. 12, 178–180 (2006).
Kelly, C. J., Zheng, L., Taylor, C. T. & Colgan, S. P. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17, 662–671 (2015).
Glover, L. E., Lee, J. S. & Colgan, S. P. Oxygen metabolism and barrier regulation in the intestinal mucosa. J. Clin. Invest. 126, 3680–3688 (2016).
Riggs, M. G., Whittaker, R. G., Neumann, J. R. & Ingram, V. M. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 268, 462–464 (1977).
Goodman, A. L. et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl Acad. Sci. USA 108, 6252–6257 (2011).
McNulty, N. P. et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med 3, 106ra106 (2011).
Rey, F. E. et al. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc. Natl Acad. Sci. USA 110, 13582–13587 (2013).
Krautkramer, K. A. et al. Diet–microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 64, 982–992 (2016).
Acknowledgements
The authors would like to thank C. Pan (UCLA) for help in 16S sequencing, G. A. Barrett-Wilt (University of Wisconsin Mass Spectrometry Facility) for technical support with GC/MS analysis, D. A. Roenneburg (Department of Surgery, University of Wisconsin School of Medicine and Public Health) for assistance with histology and B.D. Mickelson (Envigo) for assistance with diets. We also thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing sequencing and support services. This work was supported in part by grants NIH DK108259 (to F.E.R.) and HL30568 (to A.J.L.), by the National Institute of Food and Agriculture, US Department of Agriculture, under award number 2016-67017-24416 (to F.E.R.) and the Swedish Heart Lung Foundation (to F.B.). This work was also supported in part by a grant from a Transatlantic Networks of Excellence Award from the Leducq Foundation. K.K. is supported by the Astellas Foundation for Research on Metabolic Disorders, the International Atherosclerosis Society, the Yamada Science Foundation and the Sumitomo Life Welfare and Culture Foundation. K.A.K. is supported by NIH F30 DK108494-02.
Author information
Authors and Affiliations
Contributions
F.E.R. conceived, designed and supervised the project. K.K. designed the project, performed most of the experiments, analysed the results and generated figures and tables. K.A.K. performed the PTM analysis, analysed results and generated the figures. K.A.R. and K.K. performed COPRO-Seq analysis. R.L.K. cultured bacterial strains. F.B. provided GF ApoE−/− mice and E.I.V. maintained the GF mouse facility. A.J.L. and M.M. conceived and performed the Ath-HMDP experiment, and E.O. generated and analysed 16S rRNA gene data. K.K., K.A.K., R.L.K. and F.E.R. wrote the manuscript. A.J.L., M.M., F.B. and J.M.D. provided critical feedback. All authors read and agreed on the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–18
Supplementary Tables
Supplementary Tables 1–20
Rights and permissions
About this article
Cite this article
Kasahara, K., Krautkramer, K.A., Org, E. et al. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol 3, 1461–1471 (2018). https://doi.org/10.1038/s41564-018-0272-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0272-x
This article is cited by
-
Habitual carbohydrate intake is not correlated with circulating β-hydroxybutyrate levels in pregnant women with overweight and obesity at 28 weeks' gestation
Diabetologia (2024)
-
Novel primers to identify a wider diversity of butyrate-producing bacteria
World Journal of Microbiology and Biotechnology (2024)
-
Next-generation probiotics: the upcoming biotherapeutics
Molecular Biology Reports (2024)
-
Gut microbes exacerbate systemic inflammation and behavior disorders in neurologic disease CADASIL
Microbiome (2023)
-
LuQi Formula relieves ventricular remodeling through improvement of HIF-1α-mediated intestinal barrier integrity
Chinese Medicine (2023)