Abstract
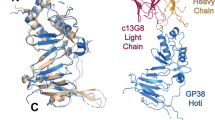
Ebola virus (EBOV) in humans causes a severe illness with high mortality rates. Several strategies have been developed in the past to treat EBOV infection, including the antibody cocktail ZMapp, which has been shown to be effective in nonhuman primate models of infection1 and has been used under compassionate-treatment protocols in humans2. ZMapp is a mixture of three chimerized murine monoclonal antibodies (mAbs)3,4,5,6 that target EBOV-specific epitopes on the surface glycoprotein7,8. However, ZMapp mAbs do not neutralize other species from the genus Ebolavirus, such as Bundibugyo virus (BDBV), Reston virus (RESTV) or Sudan virus (SUDV). Here, we describe three naturally occurring human cross-neutralizing mAbs, from BDBV survivors, that target an antigenic site in the canonical heptad repeat 2 (HR2) region near the membrane-proximal external region (MPER) of the glycoprotein. The identification of a conserved neutralizing antigenic site in the glycoprotein suggests that these mAbs could be used to design universal antibody therapeutics against diverse ebolavirus species. Furthermore, we found that immunization with a peptide comprising the HR2–MPER antigenic site elicits neutralizing antibodies in rabbits. Structural features determined by conserved residues in the antigenic site described here could inform an epitope-based vaccine design against infection caused by diverse ebolavirus species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Qiu, X. et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53 (2014).
Lyon, G. M. et al. Clinical care of two patients with Ebola virus disease in the United States. N. Engl. J. Med. 371, 2402–2409 (2014).
Qiu, X. et al. Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clin. Immunol. 141, 218–227 (2011).
Qiu, X. et al. Successful treatment of Ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci. Transl. Med. 4, 138ra81 (2012).
Olinger, G. G. Jr et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc. Natl Acad. Sci. USA 109, 18030–18035 (2012).
Wilson, J. A. et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science 287, 1664–1666 (2000).
Murin, C. D. et al. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc. Natl Acad. Sci. USA 111, 17182–17187 (2014).
Davidson, E. et al. Mechanism of binding to Ebola virus glycoprotein by the ZMapp, ZMAb, and MB-003 cocktail antibodies. J. Virol. 89, 10982–10992 (2015).
Flyak, A. I. et al. Cross-reactive and potent neutralizing antibody responses in human survivors of natural ebolavirus infection. Cell 164, 392–405 (2016).
Zhao, Y. et al. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature 535, 169–172 (2016).
Weissenhorn, W., Carfi, A., Lee, K. H., Skehel, J. J. & Wiley, D. C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2, 605–616 (1998).
Bornholdt, Z. A. et al. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science 351, 1078–1083 (2016).
Wec, A. Z. et al. Antibodies from a human survivor define sites of vulnerability for broad protection against ebolaviruses. Cell 169, 878–890 (2017).
Yu, J. S. et al. Detection of Ebola virus envelope using monoclonal and polyclonal antibodies in ELISA, surface plasmon resonance and a quartz crystal microbalance immunosensor. J. Virol. Methods 137, 219–228 (2006).
Cross, R. W. et al. The domestic ferret (Mustela putorius furo) as a lethal infection model for 3 species of Ebolavirus. J. Infect. Dis. 214, 565–569 (2016).
Kozak, R. et al. Ferrets infected with Bundibugyo virus or Ebola virus recapitulate important aspects of human filovirus disease. J. Virol. 90, 9209–9223 (2016).
Ilinykh, P. A. et al. Chimeric filoviruses for identification and characterization of monoclonal antibodies. J. Virol. 90, 3890–3901 (2016).
Malashkevich, V. N. et al. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc. Natl Acad. Sci. USA 96, 2662–2667 (1999).
Lee, J. et al. Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc. Natl Acad. Sci. USA 114, E7987–E7996 (2017).
Zhao, X. et al. Immunization-elicited broadly protective antibody reveals ebolavirus fusion loop as a site of vulnerability. Cell 169, 891–904 (2017).
Muster, T. et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67, 6642–6647 (1993).
Zwick, M. B. et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75, 10892–10905 (2001).
Montero, M., van Houten, N. E., Wang, X. & Scott, J. K. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol. Mol. Biol. Rev. 72, 54–84 (2008).
Ekiert, D. C. et al. Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 (2009).
Sui, J. et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16, 265–273 (2009).
Corti, D. et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856 (2011).
Flyak, A. I. et al. Mechanism of human antibody-mediated neutralization of Marburg virus. Cell 160, 893–903 (2015).
Lee, J. E. et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182 (2008).
Lee, J. E. et al. Techniques and tactics used in determining the structure of the trimeric ebolavirus glycoprotein. Acta Crystallogr. D Biol. Crystallogr. 65, 1162–1180 (2009).
Towner, J. S. et al. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology 332, 20–27 (2005).
Lubaki, N. M. et al. The lack of maturation of Ebola virus-infected dendritic cells results from the cooperative effect of at least two viral domains. J. Virol. 87, 7471–7485 (2013).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009).
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Davidson, E. & Doranz, B. J. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology 143, 13–20 (2014).
Bodanszky, M. & Bodanszky, A. The Practice of Peptide Synthesis 2nd edn (Springer, Berlin, 1994).
Grant, G. A. Synthetic Peptides: A User’s Guide (W.H. Freeman, New York, 1992).
Fields, G. B. & Noble, R. L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J. Pept. Protein Res. 35, 161–214 (1990).
Stuber, W., Knolle, J. & Breipohl, G. Synthesis of peptide amides by Fmoc-solid-phase peptide synthesis and acid labile anchor groups. Int J. Pept. Protein Res. 34, 215–221 (1989).
Atherton, E., Cameron, L. R. & Sheppard, R. C. Peptide synthesis: part 10. Use of pentafluorophenyl esters of fluorenylmethoxycarbonyl amino acids in solid phase peptide synthesis. Tetrahedron 44, 843–857 (1988).
Acknowledgements
This project received support from the Defense Threat Reduction Agency (grant HDTRA1-13-1-0034 to J.E.C. and A.B.), US NIH grants U19 AI109711 (to J.E.C. and A.B.), U19 AI109762 (to E.O.S. and A.B.W.), NIH contract HHSN272201400058C (to B.J.D.) and R01 AI067927 (to E.O.S.). E.O.S. is an investigator in the Pathogenesis of Infectious Disease of the Burroughs Wellcome Fund. The project was supported by NCRR grant UL1 RR024975-01 and is now at the National Center for Advancing Translational Sciences, grant 2 UL1 TR000445-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank S. Graham and L. Loerinc at Vanderbilt University for help with the expression of recombinant antibodies. We thank P. Younan at UTMB for assisting with statistical calculations. We also thank A. McNeal, S. Reddy and R. Fong for technical help with shotgun mutagenesis epitope mapping. C.D.M. is supported by a National Science Foundation Graduate Research Fellowship.
Author information
Authors and Affiliations
Contributions
A.I.F., A.B. and J.E.C. conceived the study. A.I.F., N.Kuzmina., C.D.M., C.B., E.D., P.G., P.A.I., X.S., K.H., P.R., H.T., R.L., N.Kose., H.K. and G.S. conducted the experiments. C.P.G., M.L.F., D.W.W. and E.O.S. provided reagents. A.I.F., N.Kuzmina., C.D.M., E.D., P.G., P.A.I., X.S., P.R., B.J.D., T.G.K., A.B.W., A.B. and J.E.C. analysed the data. A.I.F. and J.E.C. wrote the paper. All authors reviewed, edited and approved the paper.
Corresponding authors
Ethics declarations
Competing interests
C.B., E.D. and B.J.D. are employees of Integral Molecular. B.J.D. is a shareholder of Integral Molecular. J.E.C. is a consultant for Sanofi, and is on the Scientific Advisory Boards of PaxVax, CompuVax, GigaGen and Meissa Vaccines, is a recipient of previous unrelated research grants from Moderna and Sanofi and is founder of IDBiologics. A.I.F., P.A.I., A.B. and J.E.C. are co-inventors on a patent applied for that includes the BDBV223, BDBV317 and BDBV340 antibodies.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1, Supplementary Figures 1–4.
Rights and permissions
About this article
Cite this article
Flyak, A.I., Kuzmina, N., Murin, C.D. et al. Broadly neutralizing antibodies from human survivors target a conserved site in the Ebola virus glycoprotein HR2–MPER region. Nat Microbiol 3, 670–677 (2018). https://doi.org/10.1038/s41564-018-0157-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0157-z
This article is cited by
-
A high-throughput single-particle imaging platform for antibody characterization and a novel competition assay for therapeutic antibodies
Scientific Reports (2023)
-
Structural basis for broad coronavirus neutralization
Nature Structural & Molecular Biology (2021)
-
Inhalation monoclonal antibody therapy: a new way to treat and manage respiratory infections
Applied Microbiology and Biotechnology (2021)
-
Post-exposure immunotherapy for two ebolaviruses and Marburg virus in nonhuman primates
Nature Communications (2019)
-
Cross-reactive neutralizing human survivor monoclonal antibody BDBV223 targets the ebolavirus stalk
Nature Communications (2019)