Abstract
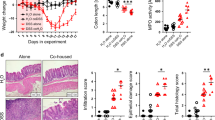
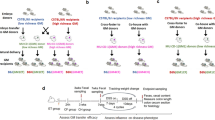
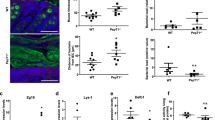
Antibiotic exposure in children has been associated with the risk of inflammatory bowel disease (IBD). Antibiotic use in children or in their pregnant mother can affect how the intestinal microbiome develops, so we asked whether the transfer of an antibiotic-perturbed microbiota from mothers to their children could affect their risk of developing IBD. Here we demonstrate that germ-free adult pregnant mice inoculated with a gut microbial community shaped by antibiotic exposure transmitted their perturbed microbiota to their offspring with high fidelity. Without any direct or continued exposure to antibiotics, this dysbiotic microbiota in the offspring remained distinct from controls for at least 21 weeks. By using both IL-10-deficient and wild-type mothers, we showed that both inoculum and genotype shape microbiota populations in the offspring. Because IL10−/− mice are genetically susceptible to colitis, we could assess the risk due to maternal transmission of an antibiotic-perturbed microbiota. We found that the IL10−/− offspring that had received the perturbed gut microbiota developed markedly increased colitis. Taken together, our findings indicate that antibiotic exposure shaping the maternal gut microbiota has effects that extend to the offspring, with both ecological and long-term disease consequences.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Jernberg, C., Löfmark, S., Edlund, C. & Jansson, J. K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66 (2007).
Cox, L. M. et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721 (2014).
Azad, M. B., Bridgman, S. L., Becker, A. B. & Kozyrskyj, A. L. Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int. J. Obesity 38, 1290–1298 (2014).
Arrieta, M.-C. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7, 307ra152 (2015).
Hviid, A., Svanstrom, H. & Frisch, M. Antibiotic use and inflammatory bowel diseases in childhood. Gut 60, 49–54 (2011).
Shaw, S. Y., Blanchard, J. F. & Bernstein, C. N. Association between the use of antibiotics and new diagnoses of Crohn’s disease and ulcerative colitis. Am. J. Gastroenterol. 106, 2133–2142 (2011).
Ng, S. C. et al. Geographical variability and environmental risk factors in inflammatory bowel disease. Gut 62, 630–649 (2013).
Kronman, M. P., Zaoutis, T. E., Haynes, K., Feng, R. & Coffin, S. E. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics 130, e794–e803 (2012).
Van Boeckel, T. P. et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect. Dis. 14, 742–750 (2014).
Hicks, L. A. et al. US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin. Infect. Dis. 60, 1308–1316 (2015).
Stokholm, J. et al. Prevalence and predictors of antibiotic administration during pregnancy and birth. PLoS ONE 8, e82932 (2013).
Andrade, S. E. et al. Prescription drug use in pregnancy. Am. J. Obstet. Gynecol. 191, 398–407 (2004).
Lacroix, I. et al. Prescription of drugs during pregnancy: a study using EFEMERIS, the new French database. Eur. J. Clin. Pharmacol. 65, 839–846 (2009).
Petersen, I., Gilbert, R., Evans, S., Ridolfi, A. & Nazareth, I. Oral antibiotic prescribing during pregnancy in primary care: UK population-based study. J. Antimicrob. Chemoth. 65, 2238–2246 (2010).
Metsälä, J. et al. Mother’s and offspring’s use of antibiotics and infant allergy to cow’s milk. Epidemiology 24, 303–309 (2013).
Moeller, A. H. et al. Cospeciation of gut microbiota. Science 353, 380–382 (2016).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Kühn, R., Löhler, J., Rennick, D., Rajewsky, K. & Müller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993).
Sellon, R. K. et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 66, 5224–5231 (1998).
Krause, P. et al. IL-10-producing intestinal macrophages prevent excessive antibacterial innate immunity by limiting IL-23 synthesis. Nat. Commun. 6, 7055 (2015).
Sartor, R. B. & Wu, G. D. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 152, 327–339 (2017).
Jones-Hall, Y. L. & Grisham, M. B. Immunopathological characterization of selected mouse models of inflammatory bowel disease: comparison to human disease. Pathophysiology 21, 267–288 (2014).
Manichanh, C. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55, 205–211 (2006).
Livanos, A. E. et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol. 1, 16140 (2016).
Pedersen, H. K. et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381 (2016).
Orešič, M. et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J. Exp. Med. 205, 2975–2984 (2008).
Nagata, C. et al. Branched-chain amino acid intake and the risk of diabetes in a Japanese community: the Takayama study. Am. J. Epidemiol. 178, 1226–1232 (2013).
Lynch, C. J. & Adams, S. H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 10, 723–736 (2014).
Attene-Ramos, M. S. et al. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ. Mol. Mutagen. 51, 304–314 (2010).
Van Passel, M. W. J. et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 6, e16876 (2011).
Cohen, S. L., Moore, A. M. & Ward, W. E. Interleukin-10 knockout mouse: a model for studying bone metabolism during intestinal inflammation. Inflamm. Bowel Dis. 10, 557–563 (2004).
Mahana, D. et al. Antibiotic perturbation of the murine gut microbiome enhances the adiposity, insulin resistance, and liver disease associated with high-fat diet. Genome Med. 8, 48 (2016).
Spencer, D. M., Veldman, G. M., Banerjee, S., Willis, J. & Levine, A. D. Distinct inflammatory mechanisms mediate early versus late colitis in mice. Gastroenterology 122, 94–105 (2002).
Berg, D. J. et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4+ Th1-like responses. J. Clin. Invest. 98, 1010–1020 (1996).
Lehmann, F. S., Burri, E. & Beglinger, C. The role and utility of faecal markers in inflammatory bowel disease. Ther. Adv. Gastroenter. 8, 23–36 (2015).
Chassaing, B. et al. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS ONE 7, e44328 (2012).
Hansen, J. J., Holt, L. & Sartor, R. B. Gene expression patterns in experimental colitis in IL-10-deficient mice. Inflamm. Bowel Dis. 15, 890–899 (2009).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Miyoshi, J. et al. Peripartum antibiotics promote gut dysbiosis, loss of immune tolerance, and inflammatory bowel disease in genetically prone offspring. Cell Rep. 20, 491–504 (2017).
Metsälä, J. et al. Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin. Exp. Allergy 45, 137–145 (2015).
Gensollen, T., Iyer, S. S., Kasper, D. L. & Blumberg, R. S. How colonization by microbiota in early life shapes the immune system. Science 352, 539–544 (2016).
Gomez de Aguero, M. et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016).
Bokulich, N. A. et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8, 343ra82 (2016).
Asnicar, F. et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2, e00164-16 (2017).
Nayfach, S., Rodriguez-Mueller, B., Garud, N. & Pollard, K. S. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 26, 1612–1625 (2016).
Palm, N. W. et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014).
Pascal, V. et al. A microbial signature for Crohn’s disease. Gut 66, 813–822 (2017).
Halfvarson, J. et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2, 17004 (2017).
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014).
Rogers, A. B. & Houghton, J. in Inflammation and Cancer: Methods in Molecular Biology Vol. 511 (ed. Kozlov, S. V.) 267–295 (Humana, New York, 2009).
Cox, D. & Snell, E. J. Analysis of Binary Data (Chapman and Hall/CRC, London, 1989).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108, 4516–4522 (2011).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Aronesty, E. Comparison of sequencing utility programs. Open Bioinformatics J. 7, 1–8 (2013).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
McDonald, D. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618 (2012).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Abubucker, S. et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8, 1002358 (2012).
Bittinger, K. qiimer: Work with QIIME Output Files in R. R package v.0.9.4 (CRAN, 2015); https://CRAN.R-project.org/package=qiimer
McDonald, D. et al. The biological observation matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. GigaScience 1, 1–6 (2012)
McMurdie, P. J. & Holmes, S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Acknowledgements
The authors thank M. Bower and the National Gnotobiotic Rodent Resource Center, University of North Carolina, Chapel Hill, for supplying mice, the NYUMC Genome Technology Center for help with sequencing (partially supported by a Cancer Center Support grant, P30CA016087, at the Laura and Isaac Perlmutter Cancer Center) and the NYUMC Histology Core for assistance preparing samples for histology. These studies were supported by NIH grants DK090989, OD010995 and DK034987 and the Crohn’s and Colitis Foundation of America, by the Ziff Family, Knapp Family and C&D funds, the Judith & Stewart Colton Center for Autoimmunity, and the Diane Belfer Program for Human Microbial Ecology.
Author information
Authors and Affiliations
Contributions
A.F.S., R.B.S. and M.J.B. designed experiments and interpreted data. A.F.S. performed experiments and participated in analysis of the data. Y.A. and V.E.R. contributed to interpretation of immunological data. Y.A. and M.H. performed protein expression assays. T.B. advised on microbiome analytical methods and performed data analyses. L.B. performed odds ratio calculations and other statistical tests. S.R., T.W. and D.K. contributed to microbiota stability analysis. A.B.R. performed histological analyses. L.M.C. and R.B.S. provided essential reagents and procedural advice. A.F.S. and M.J.B. were responsible for writing the manuscript, which was reviewed and edited by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Figures, Tables, Notes, Discussion and References.
Rights and permissions
About this article
Cite this article
Schulfer, A.F., Battaglia, T., Alvarez, Y. et al. Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat Microbiol 3, 234–242 (2018). https://doi.org/10.1038/s41564-017-0075-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-017-0075-5
This article is cited by
-
Roles of the gut microbiome in weight management
Nature Reviews Microbiology (2023)
-
Antibiotics in the pathogenesis of diabetes and inflammatory diseases of the gastrointestinal tract
Nature Reviews Gastroenterology & Hepatology (2023)
-
Analysis and Interpretation of metagenomics data: an approach
Biological Procedures Online (2022)
-
Microbiota succession throughout life from the cradle to the grave
Nature Reviews Microbiology (2022)
-
Influence of the early-life gut microbiota on the immune responses to an inhaled allergen
Mucosal Immunology (2022)