Abstract
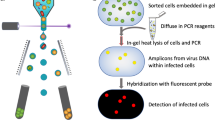
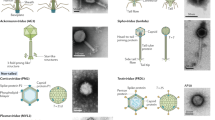
Viruses are globally abundant and extremely diverse in their genetic make-up and in the hosts they infect. Although they influence the abundance, diversity and evolution of their hosts, current methods are inadequate for gaining a quantitative understanding of their impact on these processes. Here we report the adaptation of the solid-phase single-molecule PCR polony method for the quantification of taxonomically relevant groups of diverse viruses. Using T7-like cyanophages as our model, we found the polony method to be far superior to regular quantitative PCR methods and droplet digital PCR when degenerate primers were used to encompass the group’s diversity. This method revealed that T7-like cyanophages were highly abundant in the Red Sea in spring 2013, reaching 770,000 phages ml−1, and displaying a similar depth distribution pattern to cyanobacteria. Furthermore, the abundances of two major clades within the T7-like cyanophages differed dramatically throughout the water column: clade B phages that carry the psbA photosynthesis gene and infect either Synechococcus or Prochlorococcus were at least 20-fold more abundant than clade A phages that lack psbA and infect Synechococcus hosts. Such measurements are of paramount importance for understanding virus population dynamics and the impact of viruses on different microbial taxa and for modelling viral influence on ecosystem functioning on a global scale.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bergh, O., Borsheim, K. Y., Bratbak, G. & Heldal, M. High abundance of viruses found in aquatic environments. Nature 340, 467–468 (1989).
Proctor, L. M. & Fuhrman, J. A. Viral mortality of marine bacteria and cyanobacteria. Nature 343, 60–62 (1990).
Suttle, C. A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007).
Breitbart, M. Marine viruses: truth or dare. Ann. Rev. Mar. Sci. 4, 425–448 (2012).
Güemes, A. G. C. et al. Viruses as winners in the game of life. Ann. Rev. Virol. 3, 197–214 (2016).
Zablocki, O., Adrianenssens, E. M. & Cowan, D. Diversity and ecology of viruses in hyperarid desert soils. Appl. Environ. Microb. 82, 770–777 (2016).
Breitbart, M. et al. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223 (2003).
Reyes, A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010).
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Patel, A. et al. Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescnce microscopy with SYBR Green I. Nat. Protoc. 2, 269–276 (2007).
Brussaard, C. P. D. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 70, 1506–1513 (2004).
Bratbak, G. & Heldal, M. in Handbook of Methods in Aquatic Microbial Ecology (eds Kemp, P. et al.) Ch. 16, 135–138 (CRC Press, Boca Raton, 1993).
Suttle, C. A. in Handbook of Methods in Aquatic Microbial Ecology (eds Kemp, P. et al.) Ch. 15, 121–134 (CRC Press, Boca Raton, 1993).
Kropinski, A. M., Mazzocco, A., Waddell, T. E., Lingohr, E. & Johnson, R. P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 501, 69–76 (2009).
Deng, L. et al. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513, 242–245 (2014).
Dekel-Bird, N. P., Sabehi, G., Mosevitzky, B. & Lindell, D. Host-dependent differences in abundance, composition and host range of cyanophages from the Red Sea. Environ. Microbiol. 17, 1286–1299 (2015).
Flores, C. O., Meyer, J. R., Valverde, S., Farr, L. & Weitz, J. S. Statistical structure of host-phage interactions. Proc. Natl Acad. Sci. USA 108, E288–E297 (2011).
Rozon, R. M. & Short, S. M. Complex seasonality observed amongst diverse phytoplankton viruses in the Bay of Quinte, an embayment of Lake Ontario. Freshwater Biol. 58, 2648–2663 (2013).
Tadmor, A. D., Ottesen, E. A., Leadbetter, J. R. & Phillips, R. Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science 333, 58–62 (2011).
Pinheiro, L. B. et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 84, 1003–1011 (2012).
Cai, T., Lou, G. Q., Yang, J., Xu, D. & Meng, Z. H. Development and evaluation of real-time loop-mediated isothermal amplification for hepatitis B virus DNA quantification: a new tool for HBV management. J. Clin. Virol. 41, 270–276 (2008).
Yang, H. L. et al. A novel method of real-time reverse-transcription loop-mediated isothermal amplification developed for rapid and quantitative detection of a new genotype (YHV-8) of yellow head virus. Lett. Appl. Microbiol. 63, 103–110 (2016).
Allers, E. et al. Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses. Environ. Microbiol. 15, 2306–2318 (2013).
Polz, M. F. & Cavanaugh, C. M. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64, 3724–3730 (1998).
Brankatschk, R., Bodenhausen, N., Zeyer, J. & Bürgmannc, H. Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl. Environ. Microb. 78, 4481–4489 (2012).
Brum, J. R. & Sullivan, M. B. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 13, 147–159 (2015).
Roux, S. et al. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. PeerJ 4, e2777 (2016).
Mitra, R. D. & Church, G. M. In situ localized amplification and contact replication of many individual DNA molecules. Nucl. Acids Res. 27, e34 (1999).
Sullivan, M. B., Coleman, M., Weigele, P., Rohwer, F. & Chisholm, S. W. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3, 790–806 (2005).
Labrie, S. J. et al. Genomes of marine cyanopodoviruses reveal multiple origins of diversity. Environ. Microbiol. 15, 1356–1376 (2013).
Lindell, D. et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449, 83–86 (2007).
Wang, K. & Chen, F. Prevalence of highly host-specific cyanophages in the estuarine environment. Environ. Microbiol. 10, 300–312 (2008).
Raytcheva, D. A., Haase-Pettingell, C., Piret, J. M. & King, J. A. Intracellular assembly of cyanophage Syn5 proceeds through a scaffold-containing procapsid. J. Virol. 85, 2406–2415 (2011).
Flombaum, P. et al. Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl Acad. Sci. USA 110, 9824–9829 (2013).
Dekel-Bird, N. P. et al. Diversity and evolutionary relationships of T7-like podoviruses infecting marine cyanobacteria. Environ. Microbiol. 15, 1476–1491 (2013).
Sullivan, M. B., Waterbury, J. B. & Chisholm, S. W. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424, 1047–1051 (2003).
Waterbury, J. B. & Valois, F. W. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophage abundant in seawater. Appl. Environ. Microbiol. 59, 3393–3399 (1993).
Huang, S., Zhang, S., Jiao, N. & Chen, F. Marine cyanophages demonstrate biogeographic patterns throughout the global ocean. Appl. Environ. Microbiol. 81, 441–452 (2015).
Breitbart, M., Miyake, J. H. & Rohwer, F. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol. Lett. 236, 249–256 (2004).
Labonte, J. M., Reid, K. E. & Suttle, C. A. Phylogenetic analysis indicates evolutionary diversity and environmental segregation of marine podovirus DNA polymerase gene sequences. Appl. Environ. Microbiol. 75, 3634–3640 (2009).
John, S. G. et al. A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ. Microbiol. Rep. 3, 195–202 (2011).
Matteson, A. R. et al. High abundances of cyanomyoviruses in marine ecosystems demonstrate ecological relevance. FEMS Microb. Ecol. 84, 223–234 (2013).
Lindell, D. & Post, A. F. Ultraphytoplankton succession is triggered by deep winter mixing in the Gulf of Aqaba (Eilat), Red Sea. Limnol. Oceanogr. 40, 1130–1141 (1995).
Carlson, D. F., Fredg, E. & Gildor, H. The annual cycle of vertical mixing and restratification in the Northern Gulf of Eilat/Aqaba (Red Sea) based on high temporal and vertical resolution observations. Deep Sea Res. I 84, 1–17 (2014).
Sabehi, G. et al. A novel lineage of myoviruses infecting marine cyanobacteria is widespread in the oceans. Proc. Natl Acad. Sci. USA 109, 2037–2042 (2012).
Chen, F. et al. Diverse and dynamic populations of cyanobacterial podoviruses in the Chesapeake Bay unveiled through DNA polymerase gene sequences. Environ. Microbiol. 11, 2884–2892 (2009).
Zheng, Q., Jiao, N., Zhang, R., Chen, F. & Suttle, C. A. Prevalence of psbA-containing cyanobacterial podoviruses in the ocean. Sci. Rep. 3, 3207 (2013).
Bragg, J. G. & Chisholm, S. W. Modeling the fitness consequences of a cyanophage-encoded photosynthesis gene. PLoS ONE 3, e3550 (2008).
Hellweger, F. Carrying photosynthesis genes increases ecological fitness of cyanophage in silico. Environ. Microbiol. 11, 1386–1394 (2009).
Rasband, W. S. ImageJ (US National Institutes of Health, Bethesda, Maryland, USA, 1997–2014); http://imagej.nih.gov/ij/
DeFlaun, M. F., Paul, J. H. & Jeffrey, W. H. Distribution and molecular weight of dissolved DNA in subtropical estuarine and oceanic environments. Mar. Ecol. Prog. Ser. 38, 65–73 (1987).
Kumar, S., Steher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
R Development Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria, 2013); http://www.R-project.org
Acknowledgements
We thank K. Zhang and G. Church for help with initial set-up of the polony method in our laboratory, B. Cunningham for advice on iron chloride flocculation, Lindell laboratory members for many helpful discussions and ideas during the development of the method, I. Izhaki, G. Yahel and M. Bocharenko for advice on statistical analyses, R. Kishony for use of laboratory facilities, and the Interuniversity Institute for Marine Sciences of Eilat and the Ruppin School of Marine Sciences for access to sampling facilities and B. Mosevitzky for sample collection in September 2012. We also thank S. Avrani, O. Beja, M. Breitbart, M. Carlson, Y. Mandel-Gutfreund, G. Sabehi, D. Schwartz, D. Shitrit and J. Weitz for comments on this or an earlier version of the manuscript. This research was funded by European Council FP6 Marie Curie Reintegration grant no. 046549, Israel Science Foundation Individual grant no. 749/11, European Research Council Consolidator Grant 646868 and the Mallat Family Fund and Cullen Fund from the Technion awarded to D.L.
Author information
Authors and Affiliations
Contributions
N.B. set up the polony method for viruses. N.B., S.G and I.M. designed, performed and analysed the experiments for method optimization and validation as well as field analyses, and contributed to writing of the manuscript. D.L. conceived the project, participated in experimental design and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Results and Discussion, Figures and Tables.
Rights and permissions
About this article
Cite this article
Baran, N., Goldin, S., Maidanik, I. et al. Quantification of diverse virus populations in the environment using the polony method. Nat Microbiol 3, 62–72 (2018). https://doi.org/10.1038/s41564-017-0045-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-017-0045-y
This article is cited by
-
Disentangling top-down drivers of mortality underlying diel population dynamics of Prochlorococcus in the North Pacific Subtropical Gyre
Nature Communications (2024)
-
Ubiquitous, B12-dependent virioplankton utilizing ribonucleotide-triphosphate reductase demonstrate interseasonal dynamics and associate with a diverse range of bacterial hosts in the pelagic ocean
ISME Communications (2023)
-
Abundant and cosmopolitan lineage of cyanopodoviruses lacking a DNA polymerase gene
The ISME Journal (2023)
-
Viruses affect picocyanobacterial abundance and biogeography in the North Pacific Ocean
Nature Microbiology (2022)
-
Cyanophages from a less virulent clade dominate over their sister clade in global oceans
The ISME Journal (2022)