Abstract
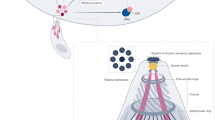
Apicomplexan parasites are important pathogens of humans and domestic animals, including Plasmodium species (the agents of malaria) and Toxoplasma gondii, which is responsible for toxoplasmosis. They replicate within the cells of their animal hosts, to which they gain access using a unique parasite-driven invasion process. At the core of the invasion machine is a structure at the interface between the invading parasite and host cell called the moving junction (MJ)1. The MJ serves as both a molecular doorway to the host cell and an anchor point enabling the parasite to engage its motility machinery to drive the penetration of the host cell2, ultimately yielding a protective vacuole3. The MJ is established through self-assembly of parasite proteins at the parasite–host interface4. However, it is unknown whether host proteins are subverted for MJ formation. Here, we show that Toxoplasma parasite rhoptry neck proteins (RON2, RON4 and RON5) cooperate to actively recruit the host CIN85, CD2AP and the ESCRT-I components ALIX and TSG101 to the MJ during invasion. We map the interactions in detail and demonstrate that the parasite mimics and subverts conserved binding interfaces with remarkable specificity. Parasite mutants unable to recruit these host proteins show inefficient host cell invasion in culture and attenuated virulence in mice. This study reveals molecular mechanisms by which parasites subvert widely conserved host machinery to force highly efficient host cell access.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Aikawa, M., Miller, L. H., Johnson, J. & Rabbege, J. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J. Cell. Biol. 77, 72–82 (1978).
Bichet, M. et al. The Toxoplasma–host cell junction is anchored to the cell cortex to sustain parasite invasive force. BMC Biol. 12, 773 (2014).
Mordue, D. G., Desai, N., Dustin, M. & Sibley, L. D. Invasion by Toxoplasma gondii establishes a moving junction that selectively excludes host cell plasma membrane proteins on the basis of their membrane anchoring. J. Exp. Med. 190, 1783–1792 (1999).
Besteiro, S., Dubremetz, J. F. & Lebrun, M. The moving junction of apicomplexan parasites: a key structure for invasion. Cell. Microbiol. 13, 797–805 (2011).
Alexander, D. L., Mital, J., Ward, G. E., Bradley, P. & Boothroyd, J. C. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 1, e17 (2005).
Besteiro, S., Michelin, A., Poncet, J., Dubremetz, J. F. & Lebrun, M. Export of a Toxoplasma gondii rhoptry neck protein complex at the host cell membrane to form the moving junction during invasion. PLoS Pathog. 5, e1000309 (2009).
Lamarque, M. et al. The RON2–AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog. 7, e1001276 (2011).
Tyler, J. S. & Boothroyd, J. C. The C-terminus of Toxoplasma RON2 provides the crucial link between AMA1 and the host-associated invasion complex. PLoS Pathog. 7, e1001282 (2011).
Tonkin, M. L. et al. Host cell invasion by apicomplexan parasites: insights from the co-structure of AMA1 with a RON2 peptide. Science 333, 463–467 (2011).
Vulliez-Le Normand, B. et al. Structural and functional insights into the malaria parasite moving junction complex. PLoS Pathog. 8, e1002755 (2012).
Beck, J. R., Chen, A. L., Kim, E. W. & Bradley, P. J. RON5 is critical for organization and function of the Toxoplasma moving junction complex. PLoS Pathog. 10, e1004025 (2014).
Lamarque, M. H. et al. Plasticity and redundancy among AMA–RON pairs ensure host cell entry of Toxoplasma parasites. Nat. Commun. 5, 4098 (2014).
Bissig, C. & Gruenberg, J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell. Biol. 24, 19–25 (2014).
Dikic, I. CIN85/CMS family of adaptor molecules. FEBS Lett. 529, 110–115 (2002).
Fisher, R. D. et al. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128, 841–852 (2007).
Kowanetz, K. et al. Identification of a novel proline–arginine motif involved in CIN85-dependent clustering of Cbl and down-regulation of epidermal growth factor receptors. J. Biol. Chem. 278, 39735–39746 (2003).
Dustin, M. L. et al. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell 94, 667–677 (1998).
Hurley, J. H. ESCRTs are everywhere. EMBO J. 34, 2398–2407 (2015).
Garrus, J. E. et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107, 55–65 (2001).
Segura-Morales, C. et al. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J. Biol. Chem. 280, 27004–27012 (2005).
Urata, S., Yokosawa, H. & Yasuda, J. Regulation of HTLV-1 Gag budding by Vps4A, Vps4B, and AIP1/Alix. Virol. J. 4, 66 (2007).
Mordue, D. G. & Sibley, L. D. A novel population of Gr-1+-activated macrophages induced during acute toxoplasmosis. J. Leukoc. Biol. 74, 1015–1025 (2003).
Schmidt, M. H., Chen, B., Randazzo, L. M. & Bogler, O. SETA/CIN85/Ruk and its binding partner AIP1 associate with diverse cytoskeletal elements, including FAKs, and modulate cell adhesion. J. Cell. Sci. 116, 2845–2855 (2003).
Campos, Y. et al. Alix-mediated assembly of the actomyosin-tight junction polarity complex preserves epithelial polarity and epithelial barrier. Nat. Commun. 7, 11876 (2016).
Johnson, R. I., Seppa, M. J. & Cagan, R. L. The Drosophila CD2AP/CIN85 orthologue Cindr regulates junctions and cytoskeleton dynamics during tissue patterning. J. Cell. Biol. 180, 1191–1204 (2008).
Gonzalez, V. et al. Host cell entry by Apicomplexa parasites requires actin polymerization in the host cell. Cell Host Microbe 5, 259–272 (2009).
Sweeney, K. R., Morrissette, N. S., LaChapelle, S. & Blader, I. J. Host cell invasion by Toxoplasma gondii is temporally regulated by the host microtubule cytoskeleton. Eukaryot. Cell 9, 1680–1689 (2010).
Tang, V. W. & Brieher, W. M. FSGS3/CD2AP is a barbed-end capping protein that stabilizes actin and strengthens adherens junctions. J. Cell. Biol. 203, 815–833 (2013).
Veiga, E. & Cossart, P. Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat. Cell Biol. 7, 894–900 (2005).
Pan, S. et al. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. J. Biol. Chem. 281, 34640–34650 (2006).
Huynh, M. H. & Carruthers, V. B. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8, 530–539 (2009).
Sheiner, L. et al. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 7, e1002392 (2011).
Sidik, S. M., Hackett, C. G., Tran, F., Westwood, N. J. & Lourido, S. Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS ONE 9, e100450 (2014).
Im, Y. J. et al. Crystallographic and functional analysis of the ESCRT-I/HIV-1 Gag PTAP interaction. Structure 18, 1536–1547 (2010).
Vojtek, A. B. & Hollenberg, S. M. Ras–Raf interaction: two-hybrid analysis. Methods Enzymol. 255, 331–342 (1995).
Fromont-Racine, M., Rain, J. C. & Legrain, P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16, 277–282 (1997).
Formstecher, E. et al. Protein interaction mapping: a Drosophila case study. Genome Res. 15, 376–384 (2005).
Wojcik, J., Boneca, I. G. & Legrain, P. Prediction, assessment and validation of protein interaction maps in bacteria. J. Mol. Biol. 323, 763–770 (2002).
Rain, J. C. et al. The protein–protein interaction map of Helicobacter pylori. Nature 409, 211–215 (2001).
London, N., Raveh, B., Cohen, E., Fathi, G. & Schueler-Furman, O. Rosetta FlexPepDock web server—high resolution modeling of peptide–protein interactions. Nucleic Acids Res. 39, W249–W253 (2011).
Raveh, B., London, N. & Schueler-Furman, O. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins 78, 2029–2040 (2010).
Crawford, J., Tonkin, M. L., Grujic, O. & Boulanger, M. J. Structural characterization of apical membrane antigen 1 (AMA1) from Toxoplasma gondii. J. Biol. Chem. 285, 15644–15652 (2010).
Lebrun, M. et al. The rhoptry neck protein RON4 relocalizes at the moving junction during Toxoplasma gondii invasion. Cell. Microbiol. 7, 1823–1833 (2005).
Lamarque, M. H. et al. Identification of a new rhoptry neck complex RON9/RON10 in the Apicomplexa parasite Toxoplasma gondii. PLoS ONE 7, e32457 (2012).
Acknowledgements
We thank P. Soubeyran for the pcDNA3-Flag-CIN85 FL plasmid, W. Sundquist for the GFP–ALIX, GST–ALIX, CHMP4B–GFP, GFP–VPS4, myc-VPS28, myc-VPS37 and myc-CD2AP plasmids, B. Striepen and M.J. Cipriano for the pU6-Cas9-YFP plasmid, P. Bradley for the anti-RON5C and S. Lourido for the pU6-universal plasmid. We are grateful to T. Soldati and B. Striepen for critical reading of the manuscript. We thank the staff of the MRI-Cytometry at the Institute for Regenerative Medicine and Biotherapy and the Flow Cytometry Facility at the Centre Médical Universitaire, Geneva, for sorting the parasites. This research was supported by the Agence Nationale de la Recherche (ANRA-12-BSV3-0012-01 and ANR-16-CE15-0010-01), Laboratoire d’Excellence (ParaFrap ANR-11-LABX-0024) and Fondation pour la Recherche Médicale (Equipe FRM DEQ20130326508) to M.L., the Canadian Institutes of Health Research grants 82915 and 148596 to M.J.B. and the Swiss National Science Foundation (FN3100A0-116722) to D.S.-F. A.G. was funded by the French Parasitology consortium ParaFrap (ANR-11-LABX-0024) and was a recipient of FRM (award no. FDT20160435387). D.J. is supported by Carigest SA. M.J.B. gratefully acknowledges the Canada Research Chairs program for salary support.
Author information
Authors and Affiliations
Contributions
M.L. conceived the study. A.G. designed, performed and interpreted most of the experimental work. R.M.C., M.H.L., H.E.H. and M.L.P. designed and performed specific experimental work. D.J. and D.S.-F. provided technical support for the cell sorting of parasites. M.L. and M.J.B. supervised the research. A.G. and M.L. wrote the paper with editorial support from M.J.B., M.H.L., D.S.-F. and D.J. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims inpublished maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Figures 1–11, full blot results, Supplementary Tables 1–3, Supplementary Text, Supplementary References.
Supplementary Table 2
List of primers.
Rights and permissions
About this article
Cite this article
Guérin, A., Corrales, R.M., Parker, M.L. et al. Efficient invasion by Toxoplasma depends on the subversion of host protein networks. Nat Microbiol 2, 1358–1366 (2017). https://doi.org/10.1038/s41564-017-0018-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-017-0018-1
This article is cited by
-
Structural insights into an atypical secretory pathway kinase crucial for Toxoplasma gondii invasion
Nature Communications (2021)
-
A lipid-binding protein mediates rhoptry discharge and invasion in Plasmodium falciparum and Toxoplasma gondii parasites
Nature Communications (2019)
-
RON4L1 is a new member of the moving junction complex in Toxoplasma gondii
Scientific Reports (2017)