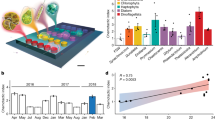
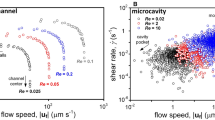
Abstract
Microbial interactions influence the productivity and biogeochemistry of the ocean, yet they occur in miniscule volumes that cannot be sampled by traditional oceanographic techniques. To investigate the behaviours of marine microorganisms at spatially relevant scales, we engineered an in situ chemotaxis assay (ISCA) based on microfluidic technology. Here, we describe the fabrication, testing and first field results of the ISCA, demonstrating its value in accessing the microbial behaviours that shape marine ecosystems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Azam, F. Microbial control of oceanic carbon flux: the plot thickens. Science 280, 694–696 (1998).
Azam, F. & Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791 (2007).
Blackburn, N., Fenchel, T. & Mitchell, J. Microscale nutrient patches in planktonic habitats shown by chemotactic bacteria. Science 282, 2254–2256 (1998).
Kiørboe, T., Grossart, H.-P., Ploug, H. & Tang, K. Mechanisms and rates of bacterial colonization of sinking aggregates. Appl. Environ. Microbiol. 68, 3996–4006 (2002).
Smriga, S., Fernandez, V. I., Mitchell, J. G. & Stocker, R. Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc. Natl Acad. Sci. USA 113, 1576–1581 (2016).
Jefferson, T. T. Zooplankton fecal pellets, marine snow and sinking phytoplankton blooms. Aquat. Microb. Ecol. 27, 57–102 (2002).
Fenchel, T. Microbial behavior in a heterogeneous world. Science 296, 1068–1071 (2002).
Adler, J. & Dahl, M. A method for measuring the motility of bacteria and for comparing random and non-random motility. Microbiology 46, 161–173 (1967).
Ford, R. M., Phillips, B. R., Quinn, J. A. & Lauffenburger, D. A. Measurement of bacterial random motility and chemotaxis coefficients: I. Stopped-flow diffusion chamber assay. Biotechnol. Bioeng. 37, 647–660 (1991).
Hol, F. J. H. & Dekker, C. Zooming in to see the bigger picture: microfluidic and nanofabrication tools to study bacteria. Science 346, 1251821 (2014).
He, K. & Bauer, C. E. Chemosensory signaling systems that control bacterial survival. Trends Microbiol. 22, 389–398 (2014).
Stocker, R. & Seymour, J. R. Ecology and physics of bacterial chemotaxis in the ocean. Microbiol. Mol. Biol. Rev. 76, 792–812 (2012).
Garren, M. et al. A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007 (2014).
Lewus, P. & Ford, R. M. Quantification of random motility and chemotaxis bacterial transport coefficients using individual-cell and population-scale assays. Biotechnol. Bioeng. 75, 292–304 (2001).
Mesibov, R. & Adler, J. Chemotaxis toward amino acids in Escherichia coli. J. Bacteriol. 112, 315–326 (1972).
Ahmed, T. & Stocker, R. Experimental verification of the behavioral foundation of bacterial transport parameters using microfluidics. Biophys. J. 95, 4481–4493 (2008).
Ammerman, J. W., Fuhrman, J. A., Hagstrom, A. & Azam, F. Bacterioplankton growth in seawater: I. Growth kinetics and cellular characteristics in seawater cultures. Mar. Ecol. Prog. Ser. 18, 31–39 (1984).
Taylor, J. R. & Stocker, R. Trade-offs of chemotactic foraging in turbulent water. Science 338, 675–679 (2012).
Stocker, R., Seymour, J. R., Samadani, A., Hunt, D. E. & Polz, M. F. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl Acad. Sci. USA 105, 4209–4214 (2008).
Azam, F. & Long, R. A. Oceanography: sea snow microcosms. Nature 414, 495–498 (2001).
Rinke, C. et al. Validation of picogram- and femtogram-input DNA libraries for microscale metagenomics. PeerJ 4, e2486 (2016).
Pedler, B. E., Aluwihare, L. I. & Azam, F. Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc. Natl Acad. Sci. USA 111, 7202–7207 (2014).
Sonnenschein, E. C. et al. Development of a genetic system for Marinobacter adhaerens HP15 involved in marine aggregate formation by interacting with diatom cells. J. Microbiol. Methods 87, 176–183 (2011).
Kelly, J. R. et al. Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Engineer. https://doi.org/10.1186/1754-1611-3-4 (2017).
Temme, K. et al. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 40, 8773–8781 (2012).
Ahmed, T., Shimizu, T. S. & Stocker, R. Microfluidics for bacterial chemotaxis. Integ. Biol. 2, 604–629 (2010).
Marie, D., Partensky, F., Jacquet, S. & Vaulot, D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR green I. Appl. Environ. Microbiol. 63, 183–186 (1997).
Parks, D. H., Tyson, G. W., Hugenholtz, P. & Beiko, R. G. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
The authors thank A. Gavish and A. Vardi for supplying V. coralliilyticus YB2, A. Stahl and M. Ullrich for supplying M. adhaerens (HP15 ΔfliC) and C. Gao and F. Moser for assistance generating the fluorescent E. coli strain used in this study. This work was supported by the Gordon and Betty Moore Foundation through a grant (GBMF3801) to J.S., G.T., P.H. and R.S. J.B.R. was supported by Australian Research Council fellowship DE160100636.
Author information
Authors and Affiliations
Contributions
B.S.L., J.-B.R., J.R.S. and R.S. designed the experiments. B.S.L., J.-B.R. and N.S. performed the experiments. B.S.L., J-B.R., V.I.F., F.R. and C.R. analysed the results. C.R., F.R., G.W.T. and P.H. generated and analysed sequencing data. B.S.L., J.-B.R., J.R.S. and R.S. wrote the manuscript. All authors edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims inpublished maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Notes 1–3 (incl. Supplementary Figures N1 and N2), Supplementary Figures 1–12 and Supplementary References.
Supplementary File 1
Computer-aided design (CAD) file for the ISCA mold; CAD file for the laboratory ISCA microcosm; CAD file for the ISCA field deployment enclosure.
Supplementary File 2
Mathematical model of chemotaxis into an ISCA well implemented in COMSOL.
Supplementary Video 1
Evolution of the chemoattractant concentration field as it diffuses from the ISCA well, as predicted by the mathematical model.
Supplementary Video 2
Accumulation of bacteria into an ISCA well predicted by the mathematical model.
Supplementary Video 3
Fluorescently labelled Vibrio coralliilyticus cells at middepth in an ISCA well containing 10% marine broth.
Supplementary Video 4
Fluorescently labelled Vibrio coralliilyticus cells at middepth in an ISCA well containing filtered artificial seawater.
Supplementary Video 5
Example of cell enumeration through image analysis in laboratory ISCA experiments, for Vibrio coralliilyticus.
Supplementary Video 6
Example of cell enumeration through image analysis in laboratory ISCA experiments, for Escherichia coli.
Supplementary Video 7
Example of cell enumeration through image analysis in laboratory ISCA experiments, for Marinobacter adhaerens.
Rights and permissions
About this article
Cite this article
Lambert, B.S., Raina, JB., Fernandez, V.I. et al. A microfluidics-based in situ chemotaxis assay to study the behaviour of aquatic microbial communities. Nat Microbiol 2, 1344–1349 (2017). https://doi.org/10.1038/s41564-017-0010-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-017-0010-9
This article is cited by
-
Chemotaxis increases metabolic exchanges between marine picophytoplankton and heterotrophic bacteria
Nature Microbiology (2023)
-
Strong chemotaxis by marine bacteria towards polysaccharides is enhanced by the abundant organosulfur compound DMSP
Nature Communications (2023)
-
Microfluidic hotspots in bacteria research: A review of soil and related advances
Soil Ecology Letters (2023)
-
The ecological roles of bacterial chemotaxis
Nature Reviews Microbiology (2022)
-
Chemotaxis may assist marine heterotrophic bacterial diazotrophs to find microzones suitable for N2 fixation in the pelagic ocean
The ISME Journal (2022)