Abstract
The surge of fast-spreading SARS-CoV-2 mutated variants highlights the need for fast, broad-spectrum strategies to counteract viral infections. In this work, we report a physical barrier against SARS-CoV-2 infection based on an inhalable bioadhesive hydrogel, named spherical hydrogel inhalation for enhanced lung defence (SHIELD). Conveniently delivered via a dry powder inhaler, SHIELD particles form a dense hydrogel network that coats the airway, enhancing the diffusional barrier properties and restricting virus penetration. SHIELD’s protective effect is first demonstrated in mice against two SARS-CoV-2 pseudo-viruses with different mutated spike proteins. Strikingly, in African green monkeys, a single SHIELD inhalation provides protection for up to 8 hours, efficiently reducing infection by the SARS-CoV-2 WA1 and B.1.617.2 (Delta) variants. Notably, SHIELD is made with food-grade materials and does not affect normal respiratory functions. This approach could offer additional protection to the population against SARS-CoV-2 and other respiratory pathogens.
Similar content being viewed by others
Main
Since its emergence in late 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused more than 600 million infections and more than 6.59 million deaths resulting from coronavirus disease 2019 (COVID-19) worldwide (as of 2 November 2022)1. Since viruses constantly change through mutation, several variants of the virus are currently variants of concern2. The emergence of new variants, especially B.1.617.2 (Delta) and B.1.1.529 (Omicron), has driven new waves of sharp increases in COVID-19 cases all over the world3. Vaccines, one of the effective strategies to protect people against infectious diseases by reducing morbidity and mortality, work primarily by producing neutralizing antibodies specific for the surface S protein4,5. However, mutation of the S protein may possibly limit the efficiency of these vaccines6,7. Thus, novel strategies that could effectively prevent infection by SARS-CoV-2, ideally before it even reaches the target lung cells, are desperately needed.
Spread through airborne transmission, SARS-CoV-2 must initially penetrate the mucus and enter the cells lining the respiratory tract8. Possible reasons for the high transmissibility of SARS-CoV-2 include an active viral replication in airway epithelia at an early stage of infection9,10. As the first and top layer of the immune system, physical barriers such as the mucus lining the airways play a key role in the defence against infections11,12. The mucus barrier offers protection in two steps: first by trapping foreign pathogens and then by eliminating them through mucociliary clearance13,14. Human airway mucus has played an important role in preventing many respiratory viruses from reaching target cells15,16; however, impaired mucociliary clearance was found for SARS-CoV-2, which promotes viral spread in the respiratory tree and increases the risk of infections17,18.
The interactions between mucus and bioadhesive polymers (such as chitosan) have been studied, opening possibilities for mucus engineering to block SARS-CoV-2 infection19. Inspired by the protective mechanism of the mucus barrier and recent advances in high-performance bioadhesives20, we designed and developed the SHIELD method to protect a subject against SARS-CoV-2. SHIELD particles are inhalable microspheres made from poly(acrylic acid) grafted with N-hydroxysuccinimide ester (PAAc-NHS ester) and gelatin. SHIELD particles interact with mucus to enhance its diffusional barrier properties, to reduce virus penetration.
Fabrication and characterization of SHIELD particles
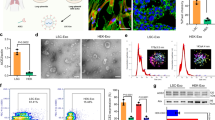
SHIELD takes effect in three steps (Fig. 1a), including inhalation (i), swelling (ii) and adhesion (iii). Preparation of the SHIELD particles was carried out using the water‐in‐oil emulsion technique. The resultant microparticles are composed of crosslinked networks of PAAc-NHS ester and gelatin (Supplementary Fig. 1), which provides a mucoadhesive with robust mechanical properties21. During the emulsion process, the physical crosslinking facilitates the transformation of the loose hydrogel structure to a dense spherical structure. Scanning electron microscopy (SEM) imaging revealed that SHIELD particles were well dispersed as a powder with a size below 5 μm (Fig. 1b). The aerodynamic diameter of the SHIELD particles was in the range of 0.5–5 μm (Fig. 1c), favouring deep lung deposition through inertial impaction and sedimentation. SHIELD particles formed a hydrogel-like structure after swelling in water (Fig. 1d). The swelling kinetics were recorded as the volume change (Fig. 1e). The volume of the SHIELD particles increased >10 times within ten minutes. A similar swelling behaviour was found in bronchoalveolar lavage fluid (Supplementary Fig. 2a,b). To enable tracking, SHIELD particles were labelled with Cyanine7 (Cy7) dye. This labelling did not affect the swelling behaviour (Supplementary Fig. 3a,b). The stability of labelled SHIELD particles was evaluated. The fluorescence signal was stable during a 72 h period (Supplementary Fig. 4). After swelling, SHIELD particles crosslinked with each other to form a hydrogel network structure (Fig. 1f). The Fourier-transform infrared spectroscopy spectrum displayed the presence of PAAc-NHS ester within the SHIELD articles (Fig. 1g). The carboxylic acid C=O stretch at 1,700 cm−1 and typical C–H stretching vibrations at 2,940 cm−1 are associated with PAAc in the SHIELD particles. The symmetric C–N–C stretch at 1,210 cm−1 and asymmetric C–N–C stretch at 1,295 cm−1 are associated with NHS ester in the SHIELD particles. The adsorption of mucin to SHIELD particles was quantified at a weight ratio of 1.14 (each milligram of SHIELD particles binds 1.14 mg of mucins; Supplementary Fig. 5). This result suggested that the mucoadhesion happens when SHIELD particles contact the wet surfaces of mucus. The hydration and swelling of SHIELD particles starts at the beginning of mucoadhesion, during which the negatively charged carboxylic acid groups in the PAAc-NHS ester can facilitate the process20,22. Simultaneously, these carboxylic acid groups form intermolecular bonds (for example, hydrogen bonds and electrostatic interactions) with the tissue surfaces23. Moreover, the NHS ester groups grafted on the PAAc can also covalently couple with the primary amine groups on tissues for further adhesion24, which was confirmed by incubating SHIELD particles with amine-coupled fluorescent beads (Fig. 1h).
a, Schematic showing the concept of the SHIELD method. Dry SHIELD particles (grey spheres) are inhaled and they become swollen (blue spheres) once they are in contact with the mucus layer (pink layer). Finally, it forms a layer of hydrogel (blue layer) and adheres to the mucus layer. The process includes inhalation (i), swelling (ii) and adhesion (iii). b, A representative SEM image showing the morphology of SHIELD particles before swelling. The inset shows a zoomed-in view of one particle. c, Aerodynamic diameter of SHIELD particles. Data are mean ± s.d.; n = 4 independent experiments. d–f, Swelling behaviour study. d, Representative optical microscopy images showing the morphological changes of SHIELD particles during a period of ten minutes in water. Scale, 50 μm. e, Volume change of spherical hydrogel in contact with water. The volume was calculated from the measured diameters during the swelling process. Data are mean ± s.d.; n = 5 independent experiments. f, A representative SEM image showing the hydrogel network after SHIELD particle swelling. g, Fourier-transform infrared spectrum showing the presence of PAAc-NHS ester within SHIELD particles. h, Mucoadhesive study using NH2-labelled beads. Fluorescence images showing the interaction between microbeads (blue) and swollen SHIELD particles (red). In b and h, independent experiments were performed (n = 5) with similar results.
SHIELD hydrogel network reinforces the mucus barrier
We hypothesized that the SHIELD network could reinforce the mucus barrier by its swelling and adhesion behaviour. Due to the limited volume of airway mucus in porcine tracheal tubes, we collected porcine stomach mucus for the test. Gastric mucins are encoded by mucin 5AC (MUC5AC) and mucin 6 (MUC6) genes, while airway mucins are encoded by MUC5AC and mucin 5B (MUC5B) genes25. MUC5AC, MUC5B and MUC6 share sequence similarities and common macromolecular characteristics, and all of them have regions rich in cysteine residues (Cys domains). Compared to mucus alone, mucus + SHIELD exhibited a denser morphology with smaller porous size (Fig. 2a), which may result from the interaction of SHIELD particles with mucus. The interaction of SHIELD particles and mucin was further observed through microscopic examination with wheat germ agglutinin staining (green; Supplementary Fig. 6) and in a three-dimensional airway model (Supplementary Fig. 7). A 1H nuclear magnetic resonance (NMR) spectrum confirmed enhanced chemical shifts between 5.5 and 8 ppm (characteristic proton in amide –CONH group)26 in mucus + SHIELD (Fig. 2b), suggesting the interaction between the carboxyl groups (on SHIELD particles) and the primary amine groups (on mucus). With a higher intensity compared to SHIELD itself, more amide bonds could potentially form between SHIELD particles and mucus. Rheology studies (Fig. 2c,d) revealed that both the elastic (G′) and viscous (G″) moduli of mucus were increased with SHIELD particles. To simulate the penetration of virus through the mucus barrier, we used polystyrene particles of the same size as virus. By tracking the trajectories of particles, we found that the addition of SHIELD particles in the mucus substantially reduced the Brownian motion of the polystyrene particles (Fig. 2e). The restricted diffusion was evaluated via the quantification of the mean square displacement (Fig. 2f). SHIELD particles reinforced the mucus barrier to decrease particle penetration (Supplementary Fig. 8). Since the transmission of SARS-CoV-2 is by inhalation of a respiratory aerosol, we used a nebulizer to generate an aerosol containing the polystyrene beads (Supplementary Fig. 9a), which are similar in size to airborne viral particles. The trajectories of the particles in the control and SHIELD groups were analysed (Supplementary Fig. 9b,c). As shown in Supplementary Fig. 9d, the motion of particles in the SHIELD group was restricted, with the mean square displacement reduced by fivefold. In addition, the morphological change of the mucus layer with SHIELD particles was observed on the surface of a porcine tracheal tube (Fig. 2g and Extended Data Fig. 1). Compared to the original mucus layer, the interaction of SHIELD particles with the mucin layer after spraying results in a denser network morphology after 10 min.
a, Optical images and SEM images showing control mucus and mucus + SHIELD after cryodesiccation. b, The 1H NMR spectra for mucus, SHIELD and mucus + SHIELD. The insets show a vertically stretched view of the region on the left side. f1, frequency dimension one. c, Frequency sweep of the elastic modulus (G′) and viscous modulus (G″) of mucus (i), mucus + SHIELD (10 mg ml–1; ii) and mucus + SHIELD (20 mg ml–1; iii). Data are mean ± s.d.; n = 3 independent experiments. d, Summary of average G′ (left) and G″ (right) at a frequency of 1.0 Hz for mucus (i), mucus + SHIELD (10 mg ml–1; ii) and mucus + SHIELD (20 mg ml–1; iii). Data are mean ± s.d.; P < 0.05 was considered statistical significance. Statistical analysis was performed with ordinary one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons, n = 3 independent experiments. e, Representative trajectories of particles within mucus and mucus + SHIELD at the same timescale. The trajectories were shifted according to their mean position. f, Ensemble-averaged geometric mean square displacements as a function of timescale; n = 48 for control group; n = 67 for SHIELD group. g, The surface morphology of a pig tracheal tube before and after a direct spray of SHIELD particles. The swelling reaction of the SHIELD particles was stopped by dipping the samples into 100% ethanol (30 s) for dehydration after 10 s or 10 min. Scale bar, 10 μm. In a and g, independent experiments were performed (n = 3) with similar results.
SHIELD inhalation blocks SARS-CoV-2 pseudo-viruses in a mouse model
Before testing in a live SARS-CoV-2 model, the protective effects of SHIELD were first tested in a mouse model with a SARS-CoV-2 pseudo-virus challenge (Fig. 3a). Inhalation in mice was performed with a modified ‘nose only’ apparatus at a dose of 3 mg SHIELD particles per kilogram of body weight (Supplementary Fig. 10). As shown in Fig. 3b, the signal of the SHIELD particles remained high in the lungs 8 h after a single inhalation but decreased from 8 h to 24 h, with minimal signal after 48 h, indicating the clearance of the SHIELD particles. A control study showed that Cy7-labelled SHIELD particles did not interfere with the luminescent/fluorescent signal in in vivo imaging system (IVIS) (Supplementary Fig. 11). As for the fluorescence signal that appeared in the liver, we speculate that the clearance is also through liver metabolization after air-to-blood translocation. In addition, mucus turnover can contribute to the clearance of SHIELD particles through the mucociliary clearance mechanism. Hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) indicated that inhaled SHIELD particles coated the surfaces of both the bronchi (Fig. 3c) and small bronchioles (Fig. 3d). To study the protection offered by SHIELD particles against variants, SARS-CoV-2 pseudo-type viruses with both D614G mutations and D614G, E484K, N501Y and K417N mutations were intranasally instilled in mice 4 h, 8 h or 24 h after SHIELD inhalation. IVIS imaging revealed that pre-inhalation of SHIELD particles blocked viral retention (baculovirus pseudotyped with D614G mutations) in the lung, with a blocking efficiency of 75.8% at 4 h, 57.7% at 8 h and 17.7% at 24 h (Fig. 3e). Confocal microscopy (Fig. 3f) and S protein IHC (Fig. 3g) confirmed that inhalation of SHIELD particles reduced the amount of SARS-CoV-2 pseudo-virus in the lung tissue. Similar protection efficiency was observed in mice challenged with baculovirus pseudotyped with part of the B.1.1.7 spike protein (D614G, E484K, N501Y and K417N mutations), showing a blocking efficiency of 71.8% at 4 h, 60.6% at 8 h and 15.1% at 24 h (Fig. 3h). This was confirmed by immunofluorescence staining (Fig. 3i) and S protein IHC (Fig. 3j). To study the broad-spectrum protection effects of SHIELD, we tested those particles against mouse pneumonia virus and H1N1 flu virus in mice. The H1N1 virus was labelled with 1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindodicarbocyanine, 4-Chlorobenzenesulfonate Salt (DiD) before challenging, which was confirmed by Fӧrster resonant energy transfer assay and the co-staining of lung sections (Supplementary Figs. 12 and 13). Our results indicated that SHIELD protection was also effective for those two pathogens (Supplementary Figs. 14 and 15).
a, A schematic showing the animal study design. Created with BioRender.com. dpi, days post-infection. b, Representative ex vivo IVIS imaging of the main organs from mice at different time points. Data are mean ± s.d.; n = 3 animals per group. The images show lung (1), heart (2), liver (3), spleen (4) and kidney (5). Corresponding quantification of fluorescence from lung is shown on the right. p, photons. c,d, H&E staining (left) and fluorescence image (right) showing the lining of a SHIELD-formed hydrogel on the airway: bronchi (c) and small bronchioles (d). Blue, 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei; red, Cy7-labelled SHIELD. Scale bars, 100 μm. Lungs were harvested from mice 4 h after inhalation. Pseudo-virus did not replicate but only delivered a genetically encoded fluorescent reporter, of which the fluorescence signal can be detected by IVIS. e,h, Representative ex vivo IVIS imaging of lungs from mice challenged with SARS-CoV-2 pseudo-virus with D614G mutation (e) and D614G, E484K, N501Y and K417N mutations (h). SHIELD particles were inhaled at 4 h, 8 h or 24 h before the challenge. The bottom graphs show the quantitative infection rates of the pseudo-virus (left) and the anti-pseudo-virus efficiency of SHIELD (right) at different time points after inhalation. The infection rate of the pseudo-virus was calculated as the radiant efficiency ((p s−1 cm–2 sr–1)/(μW cm–2)) from the infected area divided by that from the whole lung. The protective efficiency of each group was calculated as follows: ((Infection rateControl – Infection rate4h/8h/24h)/Infection rateControl) × 100. Data are shown as mean ± s.d.; n = 3 animals per group. f,i, Representative confocal images of pseudo-virus in lung tissue from mice challenged with SARS-CoV-2 pseudo-virus with D614G mutation (f) and D614G, E484K, N501Y and K417N mutations (i), 8 h after SHIELD particle inhalation. proSPC, anti-Prosurfactant Protein C. Scale bars, 50 μm. g,j, Representative images of SARS spike protein IHC staining in lung tissue from mice challenged with SARS-CoV-2 pseudo-virus with D614G mutation (g) and D614G, E484K, N501Y and K417N mutations (j), 8 h after SHIELD particle inhalation. H score, histochemical scoring. Scale bars, 50 μm. The graphs show the quantitative analysis. Data are shown as mean ± s.d.; n = 3 animals per group. Statistical analysis was performed by two-tailed unpaired t-test. In c, d, f and i, independent experiments were performed (n = 3) with similar results.
The protection of SHIELD in post-infection situations was also investigated using pneumonia virus, where mice were challenged before SHIELD inhalation. Virus shedding from the upper respiratory tract, which is regarded as a marker of infectiousness to predict the efficiency of viral transmission, were collected by nasal swab at different time points after SHIELD inhalation (4 h, 8 h and 24 h). There was no difference between the control group and the SHIELD inhalation group (Supplementary Fig. 16) in the quantification through polymerase chain reaction (PCR), indicating the minimal efficacy of SHIELD on post-infection individuals.
To study the biocompatibility of SHIELD, cells were cultured with SHIELD particles at different concentrations for three days. Even at a high concentration (10 mg ml–1), cell viability was maintained at 95% (Supplementary Fig. 17a,b). To study the toxicity of repeated SHIELD dosing, mice were treated with SHIELD particles daily for two weeks. All pulmonary function tests returned with a normal reading (80–120% of baseline), including inspiratory capacity, respiratory elastance, hysteresis area and ratio of forced expiratory volume to forced vital capacity (Supplementary Fig. 18a–d). This indicated that the inhalation of SHIELD particles did not affect normal lung functions. After two weeks of dosing, major organs were harvested for H&E staining. There was no histological evidence of injury observed compared to control animals (Supplementary Fig. 18e). Tracheas and bronchi harvested from these mice had normal morphologies (Supplementary Fig. 19), suggesting the safety of repeated administration of SHIELD particles. To determine if the inhalation of SHIELD particles affects the normal function of mucociliary clearance, mice were treated with SHIELD either acutely (2 h) or long term (daily inhalation for two weeks). The results suggested that inhalation of SHIELD particles had minimal effects on pulmonary mucociliary clearance (Supplementary Fig. 20) and did not cause long-term crosslinking of mucus. Like other reported biopolymers27, SHIELD particles only induced a temporary modification of mucus. Since the mucus layer acts not only as a physical barrier but also a biochemical barrier28, the level of secretory immunoglobulin A (sIgA) antibody in the bronchoalveolar lavage fluid was evaluated before and after interaction with SHIELD particles. Enzyme-linked immunoassay (ELISA) indicated unchanged sIgA levels after a daily SHIELD inhalation for two weeks (Supplementary Fig. 21), suggesting that mucosal immunity was not harmed by administration of the SHIELD method.
SHIELD inhalation protects African green monkeys from SARS-CoV-2 infection
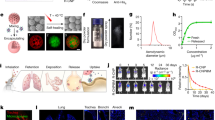
To evaluate the protection efficacy of the SHIELD method, a pilot non-human primate study was performed using both the original SARS-CoV-2 WA1 and the B.1.617.2 (Delta) variant. SHIELD inhalation was performed 8 h before the virus challenge, using either SARS-CoV-2 WA1 or the B.1.617.2 (Delta) variant, by intranasal and intratracheal routes (Fig. 4a). Nasal swabs and bronchoalveolar lavage were collected on days 1, 2, 4 and 7. A viral load assay was performed by real-time PCR with reverse transcription specific for viral subgenomic RNA (sgRNA; indicative of virus replication; Fig. 4b). Overall, animals protected with SHIELD had viral loads from 50-fold to 300-fold less than the control animals (Fig. 4c). The chemicals in SHIELD did not affect the integrity of the PCR experiment (Supplementary Fig. 22). The protection was seen with both SARS-CoV-2 WA1 and the B.1.617.2 (Delta) variant. Temperature and body weight fluctuations were not remarkable in both the control and SHIELD-protected animals (Supplementary Figs. 23 and 24). According to the literature, transient fever may be a feature of disease in non-human primate models of COVID-19, but temperature changes were not consistently observed in many cases. Viral load is the direct evidence of virus infection29. Although non-human primates are not able to develop all aspects of overt debilitating COVID-19 clinical illness as seen in humans, such as respiratory symptoms, they are still considered a gold standard model for developing new vaccines and therapeutics30,31,32.
a, Schematic depicting the non-human primate study design. Created with BioRender.com. BAL, bronchoalveolar lavage; NS, nasal swab. b, Viral sgRNA copies per swab in nasal swabs and per millilitre in bronchoalveolar lavage at various time points following challenge. Each dot represents data from one animal; n = 3 animals per group; *P < 0.05. Statistical analysis was performed by two-way ANOVA. c, Viral loads calculated based on the area under the curve (AUC) from the control or SHIELD-protected animals. Data are mean ± s.d.; n = 3 per group. d, Representative H&E images of fixed lung tissues from SARS-CoV-2-infected African green monkeys. At least seven images were taken per animal. Scale bar, 500 μm. The right side shows the quantification of lung fibrosis of the infected monkeys by Ashcroft scoring; seven tissue slices were analysed for each animal. Data are shown as mean ± s.d.; n = 21 per group; *P < 0.05. Statistical analysis was performed by two-tailed Mann–Whitney test. Ashcroft scoring was performed blindly. e, Representative images of SARS-N IHC staining in fixed lung tissues from SARS-CoV-2-infected monkeys 7 d post viral challenging; n = 3 animals per group. The arrows indicate SARS-N positive sites. Seven images were taken for each animal. Scale bar, 100 μm. f, Quantitation of positive SARS-N numbers in lung tissues of infected monkeys. Data are shown as mean ± s.d.; n = 21 per group; *P < 0.05. Statistical analysis was performed by two-tailed Mann–Whitney test. g, Representative images of RNAscope in situ hybridization detection of vRNA in infected monkeys; n = 3 animals per group. Seven images were taken for each animal. Scale bar, 100 μm. Corresponding quantifications are shown on the right; n = 21 biologically independent samples. Data are mean ± s.d.; *P < 0.05. Statistical analysis was performed by two-tailed Mann–Whitney test. h,i, Representative immunofluorescence images of SARS-N (greyscale), CD206 (green) and DAPI (blue) for WA1-challenged (h) or B.1.617.2 (Delta)-challenged (i) animals. HPF, high-power field. Scale bars, 100 μm; n = 3 animals per group. Seven images were taken for each animal. Corresponding quantifications are shown on the right; n = 21 biologically independent samples. Data are mean ± s.d.; *P < 0.05. Statistical analysis was performed by two-tailed Mann–Whitney test.
H&E staining revealed more-severe inflammation and viral pneumonia in the control animals (Fig. 4d). For the non-human primate model, the formation of fibrosis can be observed five days post infection33,34, and the pulmonary pathological change of fibrosis is a marker for lung injury by interstitial pneumonia35,36. The Ashcroft score was applied here to measure the fibrosis, which revealed that SHIELD protection decreased lung fibrosis. In addition, SARS nucleocapsid protein (SARS-N) was decreased substantially by SHIELD protection (Fig. 4e,f). To visualize SARS-CoV-2 viral RNA (vRNA) in the lung tissue, in situ RNA hybridization (RNAscope) was performed (Fig. 4g). Positive-sense vRNA was largely reduced in the SHIELD group compared to the control group, indicating limited viral replication. Furthermore, less SARS-N protein and fewer CD206+ macrophages were observed in the SHIELD group (Fig. 4h,i). As a macrophage marker, CD206 has been suggested to inhibit effector T cell function in SARS-CoV-2 (ref. 37). The decrease of CD206+ macrophages with SHIELD protection indicated a reduced virus infection. Lastly, haematology analysis assured us that SHIELD particle inhalation did not cause toxicity in the animals (Supplementary Fig. 25).
Outlook
In the present study, we demonstrated the SHIELD method offering protection against infection from SARS-CoV-2 WA1 and the B.1.617.2 (Delta) variant. Fabricated from food-grade materials, SHIELD particles are both effective and safe, and work without affecting normal lung functions or causing toxicity. Unlike vaccines, the SHIELD method enhances the diffusional barrier properties of mucus to provide physical protection against virus infection. Acting as an innate defence mechanism, the airway mucus layer plays a key role in trapping and keeping pathogens away from the host epithelial cell surface38. The main component of mucus is mucins (0.2–5% w/w), which are glycoproteins with a protein core and carbohydrate side chains39,40 To penetrate the mucus barrier, viruses have developed various strategies, such as secreting mucolytic enzymes or altering the virus’s surface properties41. Studies have reported that the SARS-CoV-2 virus infects both ciliated cells and secretory cells in the human airway, resulting in its efficient upper airway transmission8,9,10. The presence of angiotensin-converting enzyme 2 (ACE2) was confirmed in the motile cilia, and ~80% of the human respiratory epithelium (from the nasal cavity down to the lower bronchus) is densely covered with cilia42. This also provides a large surface area for SARS-CoV-2 to bind and enter host cells. Hence, it would be ideal to design a strategy to strengthen the mucosal barrier without affecting the normal mucociliary clearance properties.
There are some studies demonstrating the mucus reinforcement properties of complexing polymer molecules27,43. Low molar mass mucoadhesive polymers, such as chitosan, have shown potential in mucosal barrier enhancement and have been used to develop various vaginal and intestinal dosage formulations44. Previous studies have demonstrated that various dietary compounds also alter mucus barrier properties, such as green tea polyphenol epigallocatechin gallate45. Orally administered gelatin tannate protected the gastric mucus layer from breakdown to modulate the gut microbiota composition46,47. These studies opened up the possibilities for mucus engineering; however, targeted in vivo delivery remains challenging, especially in the pulmonary system. As shown in our study, SHIELD particles were fabricated in the form of a dry powder, with a particle size of 1–5 µm. Such a size facilitated the efficient deposition by inertial impaction and sedimentation deep into the airways and lung tissues. Taking advantage of dry powder inhalation, the SHIELD method is easy to manage for daily administration and requires only low-cost inhaler devices. The SHIELD method can serve as a supplement to face masks and/or as an alternative when face masks are not used (for example, during eating and drinking, swimming or heavy exercising, or for small children who are reluctant to wear masks).
Some limitations of our study are worth noting. One limitation is that we used only three animals in each group for each variant in the non-human primate study. This sample size is common for COVID-19 studies involving non-human primates and is enough for analysis, but a larger sample size would be needed for future studies. Our experiment results suggest a lack of efficacy when the SHIELD method is applied post infection. Since the protection mechanism of SHIELD is to reinforce the mucus structure and restrict the penetration of foreign pathogens, this result is predictable, since SHIELD’s biggest effect is blocking the entrance of the virus, not reducing the amount of virus in those already infected. As the first generation of the SHIELD method, the current method is designed only for preventative purposes, not therapeutic purposes for post-infection patients. We are now trying to optimize the formula of the SHIELD method, such as by combining it with anti-viral therapeutics. The next-generation SHIELD method will be effective after infection, which will make the method be functionally closer to the use of masks, slowing the virus spread from an infected individual to uninfected contacts.
The continued mutation of the SARS-CoV-2 virus leads to the emergence of a series of viral variants, causing new rises in reported COVID-19 cases48,49. A mutation in the S protein may impair the neutralizing epitopes, reduce vaccination efficiency and increase virus transmission50. New vaccines may be needed to combat the different variants. By contrast, the SHIELD strategy will not be limited by viral mutation. Physical protection, such as masks or other facial covering methods, has shown great effectiveness in reducing virus transmission during the pandemic. Our study takes advantage of mucus as a natural mechanical barrier against foreign pathogens. SHIELD particles reinforce the mucus structure and restrict the penetration of foreign pathogens. The broad-spectrum protection of SHIELD towards other respiratory pathogens was confirmed in our studies with inactivated H1N1 virus and pneumonia virus. We reason that when used in combination with facial covering and vaccination, SHIELD can offer effective and additional protection to the general population against COVID-19 and other respiratory pathogens.
Methods
All experiments were performed in accordance with relevant guidelines and ethical regulations approved by the North Carolina State University.
Fabrication of SHIELD particles
To prepare SHIELD particles, acrylic acid (147230, Sigma-Aldrich), acrylic acid NHS ester (AAc-NHS ester; A8060, Sigma-Aldrich) and gelatin (G1890, Sigma-Aldrich) were used. SHIELD particles were prepared using a water‐in‐oil emulsion method. One gram gelatin was dissolved in 20 ml ultrapure water with 3 ml acrylic acid. This solution was warmed to 45 °C, followed by the addition of 100 mg AAc-NHS ester. The mixture was stirred at 45 °C for 20 min and sonicated for 30 s to make sure all the AAc-NHS ester was dissolved. Due to the electrostatic interaction, acrylic acid monomer and AAc-NHS ester are bound to the gelatin chain, which provides favourable conditions for the polymerization of PAAc-NHS ester during the emulsion process. This prepared solution was added dropwise to 500 ml corn oil with 0.1 wt% Tween 80. After stirring at 3,000 r.p.m. at 55 °C for 20 min, the mixture was stirred in an ice-water bath for another 2 h. To obtain the SHIELD particles, the mixture was filtered and washed with acetone. SHIELD particles were dried in a chemical hood thoroughly and stored in a desiccator for further use.
Fluorescent labelling of SHIELD particles
Briefly, Cy7 NHS ester (Lumiprobe) was dissolved in acetone at a concentration of 0.1 mg ml–1, and SHIELD particles (1 g) were added to 10 ml dye–acetone solution. The solution was stirred at room temperature overnight, and SHIELD particles were collected through centrifugation. The collected SHIELD material was further washed three times with acetone and dried in a hood before use.
Fourier-transform infrared and 1H NMR spectroscopy
The interaction between the SHIELD particles and amino groups was revealed by Fourier-transform infrared spectroscopy (Alpha, Bruker). The 1H NMR spectroscopy was performed with a Bruker Avance NEO 600 MHz NMR instrument, equipped with a room temperature BBO Smart probe, TXI 1H–13C/15N–2H probe, variable temperature (VT) and SampleXpress automatic sample changer.
Adsorption of mucin to microspheres
Some 5 ml of mucin solution (0.5 mg ml–1) was prepared, and SHIELD particles (0.5 mg) were added. After vortexing and incubation at 37 °C for 2 h, the mixture was centrifuged at 1,375g for 5 min. To determine the concentration of mucin, periodic acid reagent was added to the supernatant (1:10). After incubation at 37 °C for 2 h, Schiff reagent was added in the same volume as the periodic acid. The optical density was measured at 555 nm using a microplate reader 30 min later. The concentration of mucin was determined using a calibration curve.
SEM imaging of SHIELD particles
Conductive tape was placed on specimen stubs. SHIELD powder was dispersed on the conductive tape to form a loose layer, and unattached powder was blown away using a bulb syringe. Samples were sputter-coated with 10 nm Au plasma (30 s) before imaging. SEM images were taken using a JEOL (JCM-7000) under acceleration voltage 15.0 and in high-voltage vacuum mode.
Rheology test
We used a Molecular Compact Rheometer (MCR302, Anton Paar) equipped with a cone plate geometry of 50 mm diameter and 1 degree (CP50-1/TG, Anton Paar). The truncation gap was set at 0.1 mm. Before the measurement, the instrument inertia was checked and the system was calibrated, as is routine. Some 2 ml of sample was placed on the temperature-controlled Peltier plate at room temperture. The geometry cone was set at the measuring position, and excess sample was trimmed. The following tests were performed in triplicate: Strain sweeps were performed at an oscillatory frequency of 1 Hz and shear strain (γ) ranging from 0.1% to 100%. Sixty data points were collected over the log-scale range of shear strain values. Frequency sweeps were performed between 0.1–50 Hz at 1% shear strain. Thirty data points were collected for each measurement.
Particle tracking
Confocal laser scanning microscopy (FluoView, Olympus) was used for particle tracking under three-dimensional visualization mode. A μ-Slide well (no. 81506, Ibidi) was filled with twenty microliters freshly collected porcine stomach mucus. Some 5 μl of latex beads (L9904, 0.1 μm mean particle size, Sigma) were then loaded gently on top. The deposition of latex beads was continuously monitored for 20 min.
For the Brownian motion analysis, 500 μl mucus or mucus + SHIELD (1 mg ml–1) was added in 12-well plates and gently stirred before being incubated for 1 h. Latex beads were diluted to ×100, and 10 μl of diluted beads were added to each well without mixing. The motion of the beads was observed and recorded for 20 s using a fluorescence microscope (Revolve, ECHO). Single particle tracking was performed with the ImageJ plugin (TrackMate). Mean square displacement was quantified using MATLAB with a per-value class (@msdanalyzer).
Cell culture
Human bronchial epithelial cells were purchased from the American Type Culture Collection (PCS-300-010) and cultured in precoated flasks with a Complete Human Epithelial Cell Medium /w kit (H6621, Cell Biologics). The three-dimensional airway model (502-3D-24, Cell Applications) was cultured according to the manufacturer’s instructions and sliced (5 μm) before staining.
Mouse studies
All studies and protocols were approved by the Institutional Animal Care and Use Committee of North Carolina State University (protocol no. 19-806-B). Male CD1 mice (aged seven weeks) were obtained from Charles River Laboratory. Cy7-labelled SHIELD particles (3 mg per kilogram of body weight) were delivered to the CD1 mice via inhalation treatment using a customized dry powder inhaler. Biodistribution of SHIELD particles in mice was studied first. Mice were euthanized at 4, 8, 24, 28, 32 and 48 hours. All major organs were collected and imaged by IVIS. Then, the organs were cryo-sectioned for histology and immunofluorescence analysis. Mice inhaled SHIELD particles 4 h, 8 h or 24 h before a challenge with SARS-CoV-2 pseudo-virus with a D614G mutated spike protein (C1120G, Montana Molecular) or SARS-CoV-2 pseudo-virus with part of a B.1.1.7 spike protein (D614G, E484K, N501Y and K417N mutations; C1122G, Montana Molecular). The dose of pseudo-virus was 100 μl per mouse. Virus was labelled with green fluorescent protein: mouse pneumonia virus (20 μl per mouse, OTV-011, Creative Biogene) or H1N1 virus (100 μl per mouse, gamma radiation inactived A/New Caledonia/20/99 strain, labelled with DiD, 23-047-299, Microbiologics). One day after challenge, all major organs were collected and evaluated by IVIS imaging. After that, organs were kept in 10% neutral buffered formalin (NBF) followed by dehydration in 10%, 20% and 30% sucrose; 5 μm lung sections were prepared.
Characterization of H1N1 virus labelled with DiD
Fӧrster resonant energy transfer assay was applied using PE-Alexa 610 (Q610) (donor; excitation wavelength, 570 nm) to label anti-hemagglutinin (anti-HA) antibody (influenza A H1N1 (A/New Caledonia/20/1999) HA antibody, rabbit PAb, 1:200, 11683-RP01, SinoBiological) on the virus surface, and DiD (acceptor; excitation wavelength, 630 nm) as a Fӧrster resonant energy transfer pair. Fӧrster resonant energy transfer occurs when two molecules are close enough that the emission of the donor can be partly used as the excitation for the acceptor51,52,53. The fluorescence emission of DiD was observed when virus particles were co-labelled with Q610 and DiD, while there was no fluorescence signal observed when DiD was excited at 570 nm (Supplementary Fig. 12), indicating that the DiD is on the surface of the virus particles close to the HA antigen.
To further identity the fluorescence-labelled virus particles, mouse lungs were harvested after challenging with DiD-labelled virus particles and stained with an antibody specific to H1N1 HA (Supplementary Fig. 13). The colocalization of DiD and viral antigen was observed using confocal microscopy, revealing DiD-labelled virus particles entering into cells.
Pulmonary function and sIgA measurements
Pulmonary function measurements were performed with the FlexiVent (SCIREQ). Prior to measurements, animals were anaesthetized with an intraperitoneal injection of ketamine and xylazine solution (2:1 ratio). The animals were intubated with a cannula. Pulmonary function baseline data were recorded before inhalation on day 0. Mice inhaled SHIELD particles (3 mg kg–1 weight) daily for two weeks. For bronchoalveolar lavage fluid collection, a catheter was inserted in the trachea of the anaesthetized mouse. Then 1 ml saline solution was instilled into the bronchioles through the catheter and retracted several times. The sIgA level was evaluated with an ELISA kit (ab157717, abcam) according to the instructions.
Pulmonary mucociliary clearance test in vivo
Mucociliary clearance was determined by the elimination of microspheres from the lungs and nose. Mice were anaesthetized and intratracheally instilled with 5 × 106 carboxylate-modified yellow-green fluorescent microspheres (2 μm, F8827, Invitrogen). Mice were euthanized immediately (for the baseline measurement) or 45 min after treatment with microspheres (for control and SHIELD groups). For the SHIELD group, mice were treated with SHIELD particles daily for two weeks or 2 h before the mucociliary clearance test. Lungs with tracheas were harvested and placed in 0.1% Tween 20 phosphate-buffered saline (PBS) solution. Tissues were homogenized by a dissociator (gentleMACS) using a preset programme. Homogenized tissue solution (10 μl) was read with an automated cell counter (Countess, Thermo Fisher). The remaining fraction of microspheres was calculated by subtracting the quantity from the baseline. Pulmonary mucociliary clearance was then calculated by subtracting the remaining fraction from 100%.
Histological analysis
For H&E staining, sections were fixed in formalin (Sigma-Aldrich) for 2 min and rinsed with running water. After staining in hematoxylin (Sigma-Aldrich) for 5 min, the sections were rinsed again. Afterward, sections were dipped in pre-prepared acid alcohol for 2 s and rinsed with sodium bicarbonate. Sections were then stained with eosin (Sigma-Aldrich) for 2 min and washed with dehydrant until excess colour was washed out.
Mucus collection and preparation
A stomach was harvested from an eight-week-old male Yorkshire pig after killing (ordered from Unit II Palmetto). The procedure complied with the ethical regulations approved by the Institute Animal Care and Use Committee of North Carolina State University. The mucus was gently scraped off the stomach epithelium and diluted 1:5 in water supplemented with 200 mM NaCl, as well as 5 mM benzamidine HCl, 1 mM 2,4′-dibromoacetophenone, 1 mM phenylmethylsulfonyl fluoride and 5 mM ethylenediaminetetraacetic acid. The pH was adjusted to 7.4 with NaOH, and the mixture was gently stirred overnight at 4 °C. The solution was then centrifuged to remove cellular and food debris, and the supernatant was used for further study. For staining, mucus + SHIELD (2:1) was dissolved in PBS and gently spread on a slide. The slide was baked at 60 °C overnight to dehydrate the liquid, fixed for 10 minutes in 3.7% buffered formalin solution and then decontaminated using 70% ethanol and 1–5% phenol solutions. After washing in water, the slides were dried in air. Wheat germ agglutinin (Alexa Fluor 488 conjugated) was used for staining. After 3 h, slides were washed with PBS for further analysis.
SEM imaging of tracheas
Porcine tracheas were harvested and cut into pieces. SHIELD particles were spread evenly on the surface of the tracheas. The swelling reaction was stopped by dipping the samples into 100% ethanol (30 s) for dehydration after 10 s or 10 min. Then, tissues were fixed in 2% buffered glutaraldehyde overnight, and then rinsed with 0.1 M HEPES buffer three times (5 minutes each). A series of concentrations of alcohol was used for dehydration as listed: 50% ethanol, two times for 10 minutes each with agitation; 70% ethanol, two times for 10 minutes each with agitation; 95% ethanol, two times for 10 minutes each with agitation; and 100% ethanol, three times for 15 minutes each with agitation. Finally, tissues were dried through chemical drying with hexamethyltiisilizane (HMDS): 100% EtOH/HMDS (2:1) for 15 minutes; 100% EtOH/HMDS (1:1) for 15 minutes; 100% EtOH/HMDS (1:2) for 15 minutes; and HMDS alone for 15 minutes, three times. Samples stayed in a chemical hood overnight to evaporate the HMDS thoroughly. Samples were mounted on specimen stubs and sputter-coated with 10 nm Au plasma before imaging. SEM images were taken using a JEOL (JCM-7000) under an acceleration voltage of 15.0 and high-voltage vacuum mode.
IHC analysis in mouse studies
For immunofluorescence, lung cryosections were placed at room temperature for 30 min and then washed with PBS for 40 min. Primary antibodies were diluted with 0.01% saponin (Sigma-Aldrich) Dako solution. The antibodies were rabbit anti-firefly luciferase antibody (1:100; ab185924, Abcam), rabbit anti-Prosurfactant Protein C antibody (1:100; ab211326, Abcam) and mouse anti-SARS-CoV/SARS-CoV-2 spike antibody (1:100; 40150-D003, SinoBiological). All cryosections were incubated with primary antibodies overnight at 4 °C. Secondary antibodies including goat anti-mouse IgG H&L (Alexa Fluor 647, Abca) and goat anti-rabbit IgG H&L (Alexa Fluor 488, Abcam) were diluted in a ratio of 1:200. After washing with PBS, samples treated with primary antibodies were incubated with those secondary antibodies at room temperature for 1.5 h. Afterward, cryosections were mounted by using Prolong Gold Mounting Media with DAPI (Life Technologies). A confocal fluorescent microscope was used to image all the samples. For SARS-S IHC staining, slides were incubated with primary mouse anti-SARS-CoV/SARS-CoV-2 spike antibody (1:100; 40150-D003, SinoBiological) overnight at 4 °C. After washing with PBS, slides were incubated with goat anti-mouse HRP secondary antibody (1:200, ab97023, Abcam) for 30 minutes and then counterstained with hematoxylin for 1 min. DAB/AEC chromogen solution was added to cover the tissue sections for 1–10 min until coloured precipitate appeared. All the images were analysed using the National Institutes of Health ImageJ software.
Non-human primate studies
Twelve African green monkeys were allocated by a counterbalance randomization based on sex and weight. All animals were housed at Bioqual. SHIELD particles (3 mg kg–1 body weight) were administered by inhalation using a customized dry powder inhaler and fitted mask, 8 h before virus challenge. Six of the monkeys were challenged with SARS-CoV-2 WA1 (SARS-CoV-2, isolate USA-WA1/2020, BEI-NR-53872, lot no. 70040665) and six monkeys were challenged with SARS-CoV-2 B.1.617.2 (Delta) variant (SARS-CoV-2, isolate hCoV-19/USA/PHC658/2021, Delta Variant BEI-NR-55612, lot no. 70045240) using the intranasal and intratracheal routes. The viral inoculum (0.5 ml) was administered dropwise into each nostril, and 1.0 ml of viral inoculum was delivered intratracheally using a French rubber catheter/feeding tube (size 10, sterile; cut 4–6 inches in length). The monkeys were inoculated with a total dose of 1.0 × 105 TCID50 SARS-CoV-2 (TCID50, 50% tissue culture infectious dose). Bronchoalveolar lavage, nasal swabs, blood, body weight and body temperature were monitored or collected throughout the study. The monkeys were necropsied on day 7 post challenge. All immunologic and virologic assays were performed blinded. All animal studies were conducted in compliance with all relevant local, state and federal regulations and were approved by the Bioqual Institutional Animal Care and Use Committee.
Histopathology and IHC in African green monkeys
After fixation in 4% paraformaldehyde for 24 hours, tissues were transferred to 70% ethanol and paraffin embedded. Tissue blocks were sectioned at 5 µm thickness. To rehydrate the tissue, slides were immersed in xylene two times for 10 minutes each. Afterward, tissues were immersed in a series of graded ethanols including 100% (two times, 10 minutes each), 95% (5 minutes), 70% (5 minutes) and 50% (5 minutes). After a water rinse, slides were stained with hematoxylin (HSS16, Sigma-Aldrich) and eosin Y (318906, Sigma-Aldrich). An optical microscope was used for analysis. For SARS-N protein IHC staining, tissue sections were rehydrated as before and then treated with antigen retrieval buffer (AP9003125, Thermo) to enable antigen retrieval. Slides were incubated with primary rabbit anti-SARS-N antibody (NB100-56576, Novus, 1:200) overnight at 4 °C and then with goat anti-rabbit HRP secondary antibody (ab6721, Abcam, 1:1,000) for 1.5 h. Finally, slides were counterstained with hematoxylin followed by bluing using 0.25% ammonia water.
Microscopic lung fibrosis was scored using the Ashcroft scale based on H&E staining, which uses a numerical scale from 0 through 8 to grade fibrosis. Quantification of RNAscope intensity was performed according to the manufacturer’s instructions (https://acdbio.com/image-analysis), using ImageJ with Color Deconvolution and Weka Classifiers. For quantification of IHC, histochemical scoring (H score) was performed to assess the interpretation of immunoreactivity. H score incorporates both the staining intensity (i) and a percentage of stained cells at each intensity level (Pi), as previously discribed54,55,56,57. The i values are 0 (no evidence of staining), 1 (weak staining), 2 (moderate staining) and 3 (strong staining). The Pi values vary from 0% to 100%. The final H score is derived from the sum of i multiplied by Pi as in the equation shown below. This score, therefore, is in the range of 0 to 300.
Subgenomic RNA viral load assay
SARS-CoV-2 E gene subgenomic RNA (sgRNA) was assessed by reverse transcription PCR. SARS-CoV-2 E gene sgRNA was cloned into a pcDNA3.1 expression plasmid and transcribed to obtain RNA for standard curve generation (AmpliCap-Max T7 High Yield Message Maker Kit, Cellscript). The standard curve was used to calculate sgRNA in copies per millilitre or per swab. For reverse transcription PCR, the collected samples were reverse-transcribed (Superscript III VILO, Invitrogen). A gene expression assay (Taqman, Thermo Fisher Scientific) was customized to target the E gene sgmRNA. The quantitative PCR was performed on a QuantStudio 6 and 7 Flex Real-Time PCR System (Applied Biosystems) according to the manufacturer’s specifications. The quantitative assay sensitivity was 50 copies per millilitre or per swab.
RNAscope in situ hybridization
Tissue slides were deparaffinized and rehydrated as previously described. The retrieval was performed according to the manufacturer’s specifications. Briefly, slides were immersed in ACD P2 retrieval buffer (ACD catalog no. 322000) at 95–98 °C for 15 min and treated with Protease Plus (ACD catalog no. 322331) at 40 °C for 30 min. SARS-CoV-2 anti-sense specific probe v-nCoV2019-S (ACD catalog no. 848561) was used to target the positive-sense vRNA. RNAscope 2.5 HD Detection Reagent RED (ACD catalog no. 322360) was used for the probe hybridization and detection.
Immunofluorescence staining of African green monkey lung sections
All the slides were pretreated as previously described, including rehydration and retrieval. Tissue slides were incubated with primary antibody (rabbit anti-SARS-N, 1:200) overnight at 4 °C. The slides were then incubated with goat anti-rabbit Alexa Fluor 594 (Abcam, ab150080, 1:500) and AF-488-CD206 (Santa Cruz Biotechnologies, sc-376108, 1:150) at room temperature for 1 h. An Olympus FluoView confocal microscope was used for imaging.
Statistics and reproducibility
All experiments were performed at least three times independently. No statistical methods were used to predetermine sample sizes, but our sample sizes were based on previous studies. Animals were randomized to treatment groups. Data acquisition and analysis were performed by investigators blinded to the groups. No data were excluded from the analyses. Results are shown as mean ± standard deviation. Comparisons between two groups were performed using the two-tailed, unpaired Student’s t-test or two-tailed Mann–Whitney test for the non-human primate study. Comparisons among more than two groups were performed using one-way ANOVA, followed by Tukey’s test. P < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Information. Additional data related to this paper may be requested from the authors. Source data are provided with this paper.
References
Johns Hopkins University & Medicine, Coronavirus Resource Center, https://coronavirus.jhu.edu/(2023).
Burki, T. Understanding variants of SARS-CoV-2. Lancet 397, 462 (2021).
Tegally, H. et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592, 438–443 (2021).
Krammer, F. SARS-CoV-2 vaccines in development. Nature 586, 516–527 (2020).
Creech, C. B., Walker, S. C. & Samuels, R. J. SARS-CoV-2 vaccines. JAMA 325, 1318–1320 (2021).
Planas, D. et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596, 276–280 (2021).
Plante, J. A. et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121 (2021).
Tang, S. et al. Aerosol transmission of SARS-CoV-2? Evidence, prevention and control. Environ. Int. 144, 106039 (2020).
Harrison, A. G., Lin, T. & Wang, P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 41, 1100–1115 (2020).
Hou, Y. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020).
Wanner, A., Salathé, M. & O’Riordan, T. G. Mucociliary clearance in the airways. Am. J. Respir. Crit. Care Med. 154, 1868–1902 (1996).
Fahy, J. V. & Dickey, B. F. Airway mucus function and dysfunction. N. Engl. J. Med. 363, 2233–2247 (2010).
Antunes, M. B., Gudis, D. A. & Cohen, N. A. Epithelium, cilia, and mucus: their importance in chronic rhinosinusitis. Immunol. Allergy Clin. 29, 631–643 (2009).
Ganesan, S., Comstock, A. T. & Sajjan, U. S. Barrier function of airway tract epithelium. Tissue Barriers 1, e24997 (2013).
Randell, S. H. & Boucher, R. C. Effective mucus clearance is essential for respiratory health. Am. J. Respir. Cell Mol. 35, 20–28 (2006).
Marjolaine, V., Elisabeth, K., Edwards, M. E. & Nicolas, R. The airway epithelium: soldier in the fight against respiratory viruses. Clin. Microbiol. Rev. 24, 210–229 (2011).
Zhu, N. et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat. Commun. 11, 3910 (2020).
Brann, D. et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 6, eabc5801 (2020).
Parhi, R. Drug delivery applications of chitin and chitosan: a review. Environ. Chem. Lett. 18, 577–594 (2020).
Yuk, H. et al. Dry double-sided tape for adhesion of wet tissues and devices. Nature 575, 169–174 (2019).
Yuk, H. et al. Tough bonding of hydrogels to diverse non-porous surfaces. Nat. Mater. 15, 190–196 (2016).
Deng, J. et al. Electrical bioadhesive interface for bioelectronics. Nat. Mater. 20, 229–236 (2021).
Mao, X., Yuk, H. & Zhao, X. Hydration and swelling of dry polymers for wet adhesion. J. Mech. Phys. Solids 137, 103863 (2020).
Wu, J. et al. An off-the-shelf bioadhesive patch for sutureless repair of gastrointestinal defects. Sci. Transl. Med. 14, eabh2857 (2022).
Yuan, S. et al. Oxidation increases mucin polymer cross-links to stiffen airway mucus gels. Sci. Transl. Med. 7, 276ra27 (2015).
Djaković, S. et al. 1′-Acetylferrocene amino acid esters and amides. A simple model for parallel β-helical peptides. Tetrahedron 70, 2330–2342 (2014).
Kootala, S. et al. Reinforcing mucus barrier properties with low molar mass chitosans. Biomacromolecules 19, 872–882 (2018).
Pelaseyed, T. et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 260, 8–20 (2014).
Muñoz-Fontela, C. et al. Animal models for COVID-19. Nature 586, 509–515 (2020).
Rahman, C. K., Sharun, K., Jose, B., Sivaprasad, M. S. & Jisna, K. S. Animal models for SARS-CoV-2 infection: a tool for vaccine and therapeutic research. Trends Biomater. Artif. Organs 34, 78–82 (2020).
Albrecht, L. et al. COVID-19 research: lessons from non-human primate models. Vaccines 9, 886 (2021).
Bi, Z., Hong, W., Yang, J., Lu, S. & Peng, X. Animal models for SARS-CoV-2 infection and pathology. MedComm 2, 548–568 (2021).
Deng, W. et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science 369, 818–823 (2020).
Shi, R. et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124 (2020).
Hemmat, N. et al. The roles of signaling pathways in SARS-CoV-2 infection; lessons learned from SARS-CoV and MERS-CoV. Arch. Virol. 166, 675–696 (2021).
Johnston, S. C. et al. Development of a coronavirus disease 2019 nonhuman primate model using airborne exposure. PLoS One 16, e0246366 (2021).
Liu, L. et al. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 4, e123158 (2019).
Carlson, T. L., Lock, J. Y. & Carrier, R. L. Engineering the mucus barrier. Annu. Rev. Biomed. Eng. 20, 197–220 (2018).
Schiller, J. L. & Lai, S. K. Tuning barrier properties of biological hydrogels. ACS Appl. Bio. Mater. 3, 2875–2890 (2020).
Lieleg, O., Lieleg, C., Bloom, J., Buck, C. B. & Ribbeck, K. Mucin biopolymers as broad-spectrum antiviral agents. Biomacromolecules 13, 1724–1732 (2012).
Janssen, W. J., Stefanski, A. L., Bochner, B. S. & Evans, C. M. Control of lung defence by mucins and macrophages: ancient defence mechanisms with modern functions. Eur. Respir. J. 48, 1201–1214 (2016).
Lee, IvanT. et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat. Commun. 11, 5453 (2020).
Kurakula, M., Gorityala, S. & Moharir, K. Recent trends in design and evaluation of chitosan-based colon targeted drug delivery systems: update 2020. J. Drug Deliv. Sci. Technol. 64, 102579 (2021).
Hamedi, H., Moradi, S., Hudson, S. M., Tonelli, A. E. & King, M. W. Chitosan based bioadhesives for biomedical applications: a review. Carbohydr. Polym. 282, 119100 (2022).
Davies, H. S. et al. Reorganisation of the salivary mucin network by dietary components: insights from green tea polyphenols. PLoS One 9, e108372 (2014).
Hanafy, N. A. N., Leporatti, S. & El-Kemary, M. A. Mucoadhesive hydrogel nanoparticles as smart biomedical drug delivery system. Appl. Sci. 9, 825–836 (2019).
Scaldaferri, F., Pizzoferrato, M., Gerardi, V., Lopetuso, L. & Gasbarrini, A. The gut barrier: new acquisitions and therapeutic approaches. J. Clin. Gastroenterol. 46, 12–17 (2012).
Korber, B. et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827 (2020).
Grubaugh, N. D., Hanage, W. P. & Rasmussen, A. L. Making sense of mutation: what D614G means for the COVID-19 pandemic remains unclear. Cell 182, 794–795 (2020).
Alina, B. et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369, 1014–1018 (2020).
Ning, B., Huang, Z. & Youngquist, B.M. et al. Liposome-mediated detection of SARS-CoV-2 RNA-positive extracellular vesicles in plasma. Nat. Nanotechnol. 16, 1039–1044 (2021).
Furukawa, H., Inaba, H. & Inoue, F. et al. Enveloped artificial viral capsids self-assembled from anionic β-annulus peptide and cationic lipid bilayer. Chem. Commun 56, 7092–7095 (2020).
Kong, H., Liu, J. & Riddle, K. et al. Non-viral gene delivery regulated by stiffness of cell adhesion substrates. Nat. Mater. 4, 460–464 (2005).
McCarty, K.S., Miller, L.S., Cox, E.B., Konrath, J. & McCarty KS, S. Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch. Path. Lab 109, 716–721 (1985).
McCarty, K.S., Szabo, E. & Flowers, J.L. et al. Use of a monoclonal antiestrogen receptor antibody in the immunohistochemical evaluation of human tumors. Cancer Res. 46, 4244–4248 (1986).
Cha, Y.J., Lee, J.S. & Kim, H.R. et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS One 9, e103333 (2014).
Yoshida, A., Tsuta, K. & Wakai, S. et al. Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung cancers. Mod Pathol. 27, 711–720 (2014).
Acknowledgements
This work was supported by grants from the National Institute of Health (HL123920, HL144002, HL146153, HL149940, HL154154, HL164998 and HL147357 to K.C.) and the American Heart Association (18TPA34230092 and 19EIA34660286 to K.C.; 21CDA855570 to K.H.). We thank the Immunology Virology Quality Assessment Center at Duke Human Vaccine Institute for performing the quantitative PCR assays.
Author information
Authors and Affiliations
Contributions
K.C. and X.M. designed the overall experiments. X.M., J.L., Z.W., D.Z., K.H., S.H. and K.P. performed the experiments. X.M. analysed the data. K.C. and X.M. wrote the article. All authors read and approved the final article. All authors provided the corresponding author with written permission to be named in the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Materials thanks Valeria Cagno, Thomas Crouzier, Hyunwoo Yuk and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The morphology of SHIELD after dehydration by 100 % ethanol (30 s).
The morphology of SHIELD after exposure to 100% ethanol was investigated under optical microscopy and SEM. There was no obvious shrinkage or reversal of swelling observed after exposure to 100% ethanol for 30 s after 10 s or 10 minutes into swelling, indicating that dehydration in 100% ethanol does not affect the gel structure. Representative optical microscopy and SEM images of SHIELD after swelling reaction for 10 s (a & b) or 10 min (c & d). Scale bar, 50 μm. e, Diameter comparison between ethanol treated and nontreated groups (after 10 s swelling). Data are mean ± SD. n = 10 for each group. Statistical analysis was performed by two-sided unpaired t test.
Supplementary information
Supplementary Information
Supplementary Figs. 1–25 and references.
Source data
Source Data Fig. 1
Statistical source data for Fig. 1c,e.
Source Data Fig. 2
Statistical source data for Fig. 2c,d.
Source Data Fig. 3
Statistical source data for Fig. 3b,e,g,h,j.
Source Data Fig. 4
Statistical source data for Fig. 4b–d,f–i.
Source Data Extended Data Fig. 1
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mei, X., Li, J., Wang, Z. et al. An inhaled bioadhesive hydrogel to shield non-human primates from SARS-CoV-2 infection. Nat. Mater. 22, 903–912 (2023). https://doi.org/10.1038/s41563-023-01475-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-023-01475-7
This article is cited by
-
Inhalable extracellular vesicle delivery of IL-12 mRNA to treat lung cancer and promote systemic immunity
Nature Nanotechnology (2024)
-
A defensive blanket against viral infection of the lungs
Nature Materials (2023)