Abstract
Tissues acquire function and shape via differentiation and morphogenesis. Both processes are driven by coordinating cellular forces and shapes at the tissue scale, but general principles governing this interplay remain to be discovered. Here we report that self-organization of myoblasts around integer topological defects, namely spirals and asters, suffices to establish complex multicellular architectures. In particular, these arrangements can trigger localized cell differentiation or, alternatively, when differentiation is inhibited, they can drive the growth of swirling protrusions. Both localized differentiation and growth of cellular vortices require specific stress patterns. By analysing the experimental velocity and orientational fields through active gel theory, we show that integer topological defects can generate force gradients that concentrate compressive stresses. We reveal these gradients by assessing spatial changes in nuclear volume and deformations of elastic pillars. We propose integer topological defects as mechanical organizing centres controlling differentiation and morphogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available in the Zenodo repository https://doi.org/10.5281/zenodo.5549501. High-resolution images and videos are available from the corresponding authors upon request.
Code availability
The MATLAB code used for the 3D-orientation analysis is available in the Zenodo repository https://doi.org/10.5281/zenodo.5680045.
Change history
16 March 2022
In the version of this article initially published, Supplementary Videos 1,2 were earlier versions of the final, accepted videos, and have been now replaced in the HTML version of the article.
References
Stooke-Vaughan, G. A. & Campàs, O. Physical control of tissue morphogenesis across scales. Curr. Opin. Genet. Dev. 51, 111–119 (2018).
de Gennes, P. G. & Prost, J. The Physics of Liquid Crystals (Clarendon Press, 1995).
Kemkemer, R., Kling, D., Kaufmann, D. & Gruler, H. Elastic properties of nematoid arrangements formed by amoeboid cells. Eur. Phys. J. E 1, 215 (2000).
Junkin, M., Leung, S. L., Whitman, S., Gregorio, C. C. & Wong, P. K. Cellular self-organization by autocatalytic alignment feedback. J. Cell Sci. 124, 4213–4220 (2011).
Duclos, G., Garcia, S., Yevick, H. G. & Silberzan, P. Perfect nematic order in confined monolayers of spindle-shaped cells. Soft Matter 10, 2346–2353 (2014).
Morales-Navarrete, H. et al. Liquid-crystal organization of liver tissue. eLife 8, e44860 (2019).
Sanchez, T., Chen, D. T. N., DeCamp, S. J., Heymann, M. & Dogic, Z. Spontaneous motion in hierarchically assembled active matter. Nature 491, 431–434 (2012).
Keber, F. C. et al. Topology and dynamics of active nematic vesicles. Science 345, 1135–1139 (2014).
Kumar, N., Zhang, R., de Pablo, J. J. & Gardel, M. L. Tunable structure and dynamics of active liquid crystals. Sci. Adv. 4, eaat7779 (2018).
Thampi, S. P., Golestanian, R. & Yeomans, J. M. Instabilities and topological defects in active nematics. Europhys. Lett. 105, 18001 (2014).
Giomi, L. Geometry and topology of turbulence in active nematics. Phys. Rev. 5, 031003 (2015).
Saw, T. B. et al. Topological defects in epithelia govern cell death and extrusion. Nature 544, 212–216 (2017).
Kawaguchi, K., Kageyama, R. & Sano, M. Topological defects control collective dynamics in neural progenitor cell cultures. Nature 545, 327–331 (2017).
Duclos, G., Erlenkamper, C., Joanny, J.-F. & Silberzan, P. Topological defects in confined populations of spindle-shaped cells. Nat. Phys. 13, 58–62 (2017).
Thompson, D. W. On Growth and Form (Cambridge Univ. Press, 1942).
Mandal, S., Mahajan, D., Roy, S., Singh, M. & Khurana, N. Fibroma with minor sex cord elements: an incidental finding in a normal sized ovary—a case report with literature review. Diagn. Pathol. 2, 46 (2007).
Kepes, J. J. Cellular whorls in brain tumors other than meningiomas. Cancer 37, 2232–2237 (1976).
Dua, H. S. et al. Vortex or whorl formation of cultured human corneal epithelial cells induced by magnetic fields. Eye 10, 447–450 (1996).
Maroudas-Sacks, Y. et al. Topological defects in the nematic order of actin fibres as organization centres of Hydra morphogenesis. Nat. Phys. 17, 251–259 (2021).
Turiv, T. et al. Topology control of human fibroblast cells monolayer by liquid crystal elastomer. Sci. Adv. 6, eaaz6485 (2020).
Endresen, K. D., Kim, M., Pittman, M., Chen, Y. & Serra, F. Topological defects of integer charge in cell monolayers. Soft Matter 17, 5878–5887 (2021).
Yaffe, D. & Feldman, M. The formation of hybrid multinucleated muscle fibers from myoblasts of different genetic origin. Dev. Biol. 11, 300–317 (1965).
Duclos, G. et al. Spontaneous shear flow in confined cellular nematics. Nat. Phys. 14, 728–732 (2018).
Wioland, H., Woodhouse, F. G., Dunkel, J., Kessler, J. O. & Goldstein, R. E. Confinement stabilizes a bacterial suspension into a spiral vortex. Phys. Rev. Lett. 110, 268102 (2013).
Guillamat, P., Ignés-Mullol, J. & Sagués, F. Taming active turbulence with patterned soft interfaces. Nat. Commun. 8, 564 (2017).
Opathalage, A. et al. Self-organized dynamics and the transition to turbulence of confined active nematics. Proc. Natl Acad. Sci. USA 116, 4788–4797 (2019).
Jalal, S. et al. Actin cytoskeleton self-organization in single epithelial cells and fibroblasts under isotropic confinement. J. Cell Sci. 132, jcs220780 (2019).
Kruse, K., Joanny, J. F., Jülicher, F., Prost, J. & Sekimoto, K. Asters, vortices, and rotating spirals in active gels of polar filaments. Phys. Rev. Lett. 92, 078101 (2004).
Püspöki, Z., Storath, M., Sage, D. & Unser, M. in Focus on Bio-Image Informatics (eds De Vos, W. H., Munck, S. & Timmermans, J.-P.) 69–93 (Springer, 2016).
Tseng, Q. et al. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc. Natl Acad. Sci. USA 109, 1506–1511 (2012).
Blanch-Mercader, C., Guillamat, P., Roux, A. & Kruse, K. Quantifying material properties of cell monolayers by analyzing integer topological defects. Phys. Rev. Lett. 126, 028101 (2021).
Blanch-Mercader, C., Guillamat, P., Roux, A. & Kruse, K. Integer topological defects of cell monolayers: mechanics and flows. Phys. Rev. E. 103, 012405 (2021).
Schaller, V., Weber, C., Semmrich, C., Frey, E. & Bausch, A. R. Polar patterns of driven filaments. Nature 467, 73–77 (2010).
Lushi, E., Wioland, H. & Goldstein, R. E. Fluid flows created by swimming bacteria drive self-organization in confined suspensions. Proc. Natl Acad. Sci. USA 111, 9733–9738 (2014).
Doxzen, K. et al. Guidance of collective cell migration by substrate geometry. Integr. Biol. 5, 1026 (2013).
Fürthauer, S., Neef, M., Grill, S. W., Kruse, K. & Jülicher, F. The Taylor–Couette motor: spontaneous flows of active polar fluids between two coaxial cylinders. New J. Phys. 14, 023001 (2012).
Hobson, C. M. et al. Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Mol. Biol. Cell 31, 1788–1801 (2020).
Collinsworth, A. M., Zhang, S., Kraus, W. E. & Truskey, G. A. Apparent elastic modulus and hysteresis of skeletal muscle cells throughout differentiation. Am. J. Physiol. Cell Physiol. 283, C1219–C1227 (2002).
Da Costa, A. D. S. et al. Fibroblasts close a void in free space by a purse-string mechanism. Preprint at bioRxiv https://doi.org/10.1101/2020.08.15.250811 (2020).
Barbazan, J. et al. Cancer-associated fibroblasts actively compress cancer cells and modulate mechanotransduction. Preprint at bioRxiv https://doi.org/10.1101/2021.04.05.438443 (2021).
Gilbert, S. F. & Barresi, M. J. F. Developmental Biology 11th edn (Sinauer, 2016).
Sharples, A. P., Al-Shanti, N., Lewis, M. P. & Stewart, C. E. Reduction of myoblast differentiation following multiple population doublings in mouse C2C12 cells: a model to investigate ageing? J. Cell. Biochem. 112, 3773–3785 (2011).
Poujade, M. et al. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl Acad. Sci. USA 104, 15988–15993 (2007).
Dekkers, J. F. et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 14, 1756–1771 (2019).
Strale, P. O. et al. Multiprotein printing by light-induced molecular adsorption. Adv. Mater. 28, 2024–2029 (2016).
Huterer, D. & Vachaspati, T. Distribution of singularities in the cosmic microwave background polarization. Phys. Rev. D. 72, 043004 (2005).
Barbier de Reuille, P. et al. MorphoGraphX: a platform for quantifying morphogenesis in 4D. eLife 4, e05864 (2015).
Guo, M. et al. Cell volume change through water efflux impacts cell stiffness and stem cell fate. Proc. Natl Acad. Sci. USA 114, E8618–E8627 (2017).
Kloxin, A. M., Kloxin, C. J., Bowman, C. N. & Anseth, K. S. Mechanical properties of cellularly responsive hydrogels and their experimental determination. Adv. Mater. 22, 3484–3494 (2010).
Acknowledgements
We thank C. Roffay for suggesting the representation of 3D orientational fields and all the members from the Roux and Kruse groups for fruitful discussions and support. We thank S. Gabriele for useful insights on the elastic properties of C2C12 cell nuclei. P.G. acknowledges support from the Human Frontiers of Science Program (grant number LT-000793/2018-C). A.R. acknowledges funding from SystemsX RTD program EpiPhysX, the Swiss National Fund for Research grant numbers 31003A_130520, 31003A_149975 and 31003A_173087, and the European Research Council Consolidator grant number 311536.
Author information
Authors and Affiliations
Contributions
P.G. and A.R. designed the research. P.G. performed the experiments. P.G. and C.B.-M. analysed the data. G.P. analysed cell number density in mounds. C.B.-M. and K.K. developed the theoretical model. All the authors participated in writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Materials thanks Marino Arroyo, Michael Murrell and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Formation of a nematic cellular monolayer.
a, Time series of a proliferating monolayer of C2C12 myocytes. Scale bar, 500μm. b, Average orientational order parameter as function of time. c, Two-point spatial autocorrelation function Cnn and nematic autocorrelation length ξnn (inset). Colors correspond to the time points indicated in the inset. Data are presented as mean values + /- s.d.
Extended Data Fig. 2 Cell number density for spirals and asters.
a, Time evolution of confined cell monolayers with different initial cell number. Scale bar, 100μm. b, Time of maximum tangential speed with respect of initial cell number. Colormap indicates speed. Dashed line corresponds to a linear fit. c, Average cell number density and d, radial density profiles for spirals (N = 10) and asters (N = 9). Data are presented as mean values ± s.e.m.
Extended Data Fig. 3 Orientational field from cell-shape and actin fibers in spirals.
a, Phase contrast image of a spiral. Yellow lines correspond to the local orientation. Vector length corresponds to the coherency (see Methods). For clarity, the totality of vectors is only shown in the inset. b, Confocal micrograph of the bottom plane of an F-actin-labelled spiral. Actin was stained with SiR-actin. Scale bars, 25μm. c and d, Distributions of the angle ψ extracted from phase contrast images (N = 11) and confocal micrographs (N = 7), respectively. The angle ψ is between the local director and the radial direction with respect to the geometrical center. In panels C and D, the red dashed line indicates ψ = 90°. e and f, Radial profiles of the orientational order parameter S extracted from phase contrast images (N = 11) and confocal micrographs (N = 7), respectively. Data are presented as mean values ± s.e.m.
Extended Data Fig. 4 Influence of myosin activity on spiral morphology and dynamics.
a, Phase contrast image of a Y-27632-treated spiral. The confinement radius is 100μm. b, Average flow field for Y-27632-treated spirals. Streamlines are shown as black curves. The color code indicates the speed. c, Distributions of the angle ψ for spirals under different conditions. ψ is the angle between the local director and the radial direction with respect to the geometrical center. Dashed red line indicates ψ = 90°. d, Radial profiles of the azimuthal and e, radial velocity components for spirals under the conditions corresponding to panel C. N = 11, 20 and 21, for control spirals, Y-27632-treated spirals, and Blebbistatin-treated spirals, respectively. Data are presented as mean values ± s.e.m.
Extended Data Fig. 5 Actin flows in asters.
a, Confocal micrographs of actin at different aster bases. b, Average orientational field from time-lapses of asters in A (N = 3). Actin was stained with SiR-actin. Vector length corresponds to the coherency, showed as well as a colormap (see Methods). Scale bar, 50μm. c, Distribution of the angle ψ extracted from confocal micrographs (N = 3). The angle ψ is between the local director and the radial direction with respect to the geometrical center. Dashed red line indicates ψ = 90° (N = 3). d, Average flow field. Streamlines and vectors (inset) indicate the direction of actin flows. The colormap indicates the speed. e, Radial profiles of the radial (red) and azimuthal (black) velocity components. Data are presented as mean values ± s.e.m.
Extended Data Fig. 6 Aster arrangements and rings at the pillars’ base.
a, Time-series of a C2C12 cell monolayer initially positioned on a circular ring constricting a hydrogel micro-pillar. Cyan dashed line indicates the initial pillar section. A schematic of the setting is included as an inset. b, Actin-stained cells after constriction show an aster arrangement. Actin was stained with SiR-actin. c, Average flow field around pillars during constriction (N = 9). The color scale corresponds to the speed. d, Radial profiles of the radial (red) and azimuthal (black) velocity components. Data are presented as mean values ± s.e.m. e, Average orientational field. For clarity, only half of the total number of orientation vectors are shown. The color scale represents the angle of the director with respect to the horizontal axis. Dashed circles mark the position of the pillar section and the initial ring. Vector length corresponds to the coherency (see Methods). f, Distribution of the angle ψ between the local director and the radial direction with respect to the geometrical center. Scale bars, 50μm.
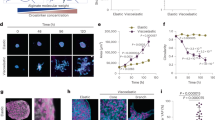
Extended Data Fig. 7 Compression dynamics of hydrogel pillars.
a, Schematics of the protocol employed to fabricate cell-adhesive rings enclosing passive fluorescent hydrogel micro-pillars. After activation of the glass substrate, micro-pillars were fabricated by illuminating a photo-polymerizable mPEG solution with disk patterns of UV light with an inverted microscope (step I). Then, the substrate was functionalized with PLL-PEG (step II). PEG chains were locally photo-degraded by illuminating the substrate with ring patterns of UV light (step III). Finally, fibronectin was incubated (step IV). b, Time-series showing the compression of a pillar. Green dashed line on bright-field images depicts the initial size of the pillar section close to the base with a radius of r = 40μm. The radius of the cellular island is 100μm. 3D rendering of the pillar is shown below. c, Temporal evolution of the height up to which pillars are compressed for different radii. d, Compressive stress for pillars with different radii. Data are presented as mean values ± s.e.m. e, Maximum compressive stress vs pillar radii. For panels C, D and E, the radius of pillars (r) is indicated in the legend of panel E. f, Maximum Z projections of a confocal stack of a mound deforming a centred pillar. g, Position of the centroid of the sections of 2 centered pillars (Green and blue). Green curves correspond to the pillar in F. Red curves correspond to a pillar without cells around it. h, Maximum Z projection of a confocal stack of a mound deforming a non-centered pillar. i, Position of the centroid of the sections of 3 non-centered pillars. Green curves correspond to the pillar in H. For G and I, centroids of the pillar base are positioned at the origin. Data are presented as mean values ± s.d. Actin was stained with SiR-actin. Nuclei were stained with Hoechst 33342. Scale bars, 50μm.
Extended Data Fig. 8 Localization of myosin heavy chain expression.
a, Image composites of phase contrast and fluorescence images of confluent monolayers of C2C12 cells competent for differentiation, after 6 days under starvation conditions (see Methods). Right panel corresponds to a zoomed region from the left panel. Scale bars, 500μm. b, Average projection of confocal micrographs of cellular islands (N = 40, base radius of 100μm) grown for 6 days in complete medium (10% FBS). Individual micrographs are shown in the panels on the right. c, Average projection of confocal micrographs of cellular islands (N = 40, base radius of 100 μm) grown for 6 days under starvation conditions (2% HS). Individual micrographs are shown in the panels on the right. For all panels, actin is shown in white, and myosin heavy chain (MyHC) is shown in yellow. Actin was stained with SiR-actin. MyHC was stained with the Myosin-4 Monoclonal Antibody conjugated with Alexa Fluor 488. Scale bars, 50μm. d, Radial profiles of averaged MyHC intensity and e, compressive stress under normal (orange) and starvation (black) conditions. Compressive stresses were measured from nuclear volume deformations. Data are presented as mean values ± s.e.m.
Extended Data Fig. 9 Cellular protrusions.
a, Image composites of phase contrast and fluorescence images of confluent monolayers of C2C12 cells incompetent for differentiation, after 6 days under starvation conditions (see Methods). Right panel corresponds to a zoomed region from the left panel. Scale bars, 500μm. MyHC was stained with the Myosin-4 Monoclonal Antibody conjugated with Alexa Fluor 488. b, Z-projections of actin of 14- (above yellow dashed line) and 16- (below yellow dashed line) days-old cellular protrusions. Red frame corresponds to a zoomed protrusion. Actin was stained with SiR-actin. Scale bars, 50μm.
Extended Data Fig. 10 Growth of cellular mounds and protrusions.
a, Average height of C2C12 islands of radius 100 µm for different time points after the onset of confluence. White symbols (○) correspond to islands of differentiation-competent cells (N = 17, 10, 16, for mounds at 1, 3, 6 days after the onset of confluence, respectively). Black symbols (●) correspond to islands of differentiation-incompetent cells (N = 42, 11, 10 and 27, for mounds at 5, 9, 12, and 16 days after the onset of confluence, respectively). Data are presented as mean values ± s.d. Inset: scheme of the time evolution of cellular nematic architectures from 2D to 3D. b, XZ section of mounds under different confinement sizes. The number of mounds in the images is 6, 5, 2 and 1 for diameters of 200, 500, 1000 and 4000 µm, respectively. Mounds were labelled with Sir Actin. c, Z projection of an actin labelled 6 day old mound with a diameter of 500 µm. Colormap indicates height. d, Aspect ratio of the mounds, calculated as height/diameter, for the different confinement sizes (N = 30, 16, 4 and 5 for diameters of 200, 500, 1000 and 4000 µm, respectively). Data are presented as mean values ± s.d. Scale bars, 200 µm.
Supplementary information
Supplementary Information
Captions for Supplementary Video 1–10, Supplementary Notes 1–8, Figs. 1–5 and References.
Supplementary Video 1
Spiral-to-aster transition in a C2C12 myoblast disk. Phase contrast time-lapse of myoblast monolayers under circular confinement. In time, cells rearrange from spiral arrangements into aster arrangements.
Supplementary Video 2
Aster-to-spiral transition in an expanding C2C12 myoblast disk. Phase contrast time-lapse of an expanding myoblast disc, from 150 to 300 μm in diameter. Blue shaded area depicts the new fibronectin-coated region. White dashed lines indicate the outer boundary of the new circular domain.
Supplementary Video 3
Formation of cellular mounds. Phase-contrast time-lapse showing the formation of cellular mounds in the centre of an aster of myoblasts.
Supplementary Video 4
Cellular spirals. Phase-contrast time-lapse of low-density circular islands of myoblasts featuring spiral configurations. Division was blocked with Mitomycin-C.
Supplementary Video 5
Actin dynamics in cellular spirals. Fluorescence confocal time-lapse of the bottom plane of a cellular spiral. Actin was stained with SiR-actin.
Supplementary Video 6
Cellular asters. Phase-contrast time-lapse of high-density circular islands of myoblasts. Division was not blocked.
Supplementary Video 7
Actin dynamics in cellular asters. Fluorescence confocal time-lapse of the bottom plane of a cellular aster. Actin was stained with SiR-actin.
Supplementary Video 8
Actin organization and flows in minimal cellular mounds. Z projection of the top of an actin-labelled mound. Actin was stained with SiR-actin.
Supplementary Video 9
Pillar constriction experiment. Differential interference contrast time-lapse showing myoblasts constricting soft hydrogel pillars of different sizes. The consequent pillars’ deformation can be observed in the 3D renderings of the pillars obtained from fluorescence images’ segmentation.
Supplementary Video 10
Dynamics and collapse of 3D cellular protrusions. Phase-contrast time-lapse of a myoblast protrusion, which collapses after confining pattern is degraded.
Rights and permissions
About this article
Cite this article
Guillamat, P., Blanch-Mercader, C., Pernollet, G. et al. Integer topological defects organize stresses driving tissue morphogenesis. Nat. Mater. 21, 588–597 (2022). https://doi.org/10.1038/s41563-022-01194-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-022-01194-5
This article is cited by
-
Defects show self-constraint in active nematics
Nature Physics (2024)
-
How multiscale curvature couples forces to cellular functions
Nature Reviews Physics (2024)
-
Hexanematic crossover in epithelial monolayers depends on cell adhesion and cell density
Nature Communications (2023)
-
Synchronization in collectively moving inanimate and living active matter
Nature Communications (2023)
-
Epithelia are multiscale active liquid crystals
Nature Physics (2023)