Abstract
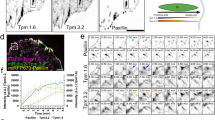
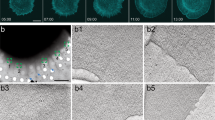
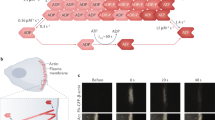
The actin cytoskeleton is the primary driver of cellular adhesion and mechanosensing due to its ability to generate force and sense the stiffness of the environment. At the cell’s leading edge, severing of the protruding Arp2/3 actin network generates a specific actin/tropomyosin (Tpm) filament population that controls lamellipodial persistence. The interaction between these filaments and adhesion to the environment is unknown. Using cellular cryo-electron tomography we resolve the ultrastructure of the Tpm/actin copolymers and show that they specifically anchor to nascent adhesions and are essential for focal adhesion assembly. Re-expression of Tpm1.8/1.9 in transformed and cancer cells is sufficient to restore cell–substrate adhesions. We demonstrate that knock-out of Tpm1.8/1.9 disrupts the formation of dorsal actin bundles, hindering the recruitment of α-actinin and non-muscle myosin IIa, critical mechanosensors. This loss causes a force-generation and proliferation defect that is notably reversed when cells are grown on soft surfaces. We conclude that Tpm1.8/1.9 suppress the metastatic phenotype, which may explain why transformed cells naturally downregulate this Tpm subset during malignant transformation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that all the data supporting the findings of this study are available within the article and the Supplementary Information. Requests for resources including plasmids should be directed to and will be fulfilled by the corresponding author. The publicly available mRNA sequence of mouse Tpm1.8/1.9 used in this study has the access code NCBI M_001164253.1. Source data are provided with this paper.
References
Gumbiner, B. M. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84, 345–357 (1996).
Zamir, E. & Geiger, B. Molecular complexity and dynamics of cell–matrix adhesions. J. Cell Sci. 114, 3583–3590 (2001).
Takada, Y., Ye, X. & Simon, S. The integrins. Genome Biol. 8, 215 (2007).
Zaidel-Bar, R., Ballestrem, C., Kam, Z. & Geiger, B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116, 4605–4613 (2003).
Zaidel-Bar, R., Cohen, M., Addadi, L. & Geiger, B. Hierarchical assembly of cell–matrix adhesion complexes. Biochem. Soc. Trans. 32, 416–420 (2004).
Wolfenson, H. et al. Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices. Nat. Cell Biol. 18, 33–42 (2016).
Meacci, G. et al. α-Actinin links extracellular matrix rigidity-sensing contractile units with periodic cell-edge retractions. Mol. Biol. Cell 27, 3471–3479 (2016).
Ghassemi, S. et al. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc. Natl Acad. Sci. USA 109, 5328–5333 (2012).
Zaidel-Bar, R., Milo, R., Kam, Z. & Geiger, B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell–matrix adhesions. J. Cell Sci. 120, 137–148 (2006).
Nakamura, K. et al. Tyrosine phosphorylation of paxillin α is involved in temporospatial regulation of paxillin-containing focal adhesion formation and F-actin organization in motile cells. J. Biol. Chem. 275, 27155–27164 (2000).
López-Colomé, A. M., Lee-Rivera, I., Benavides-Hidalgo, R. & López, E. Paxillin: a crossroad in pathological cell migration. J. Hematol. Oncol. https://doi.org/10.1186/s13045-017-0418-y (2017).
Bershadsky, A., Kozlov, M. & Geiger, B. Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr. Opin. Cell Biol. 18, 472–481 (2006).
Pellegrin, S. P. & Mellor, H. Actin stress fibres. J. Cell Sci. 120, 3491–3499 (2007).
Yang, B. et al. Stopping transformed cancer cell growth by rigidity sensing. Nat. Mater. 19, 239–250 (2020).
Sheetz, M. A tale of two states: normal and transformed, with and without rigidity sensing. Annu. Rev. Cell Dev. Biol. 35, 169–190 (2019).
Giannone, G. et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell 128, 561–575 (2007).
Swaney, K. F. & Li, R. Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr. Opin. Cell Biol. 42, 63–72 (2016).
Suraneni, P. et al. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 197, 239–251 (2012).
Brayford, S. et al. Tropomyosin promotes lamellipodial persistence by collaborating with Arp2/3 at the leading edge. Curr. Biol. 26, 1312–1318 (2016).
Bareja, I. et al. Dynamics of Tpm1.8 domains on actin filaments with single-molecule resolution. Mol. Biol. Cell 31, 2452–2462 (2020).
Tojkander, S. et al. A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol. 21, 539–550 (2011).
Bach, C. T. et al. Tropomyosin isoform expression regulates the transition of adhesions to determine cell speed and direction. Mol. Cell. Biol. 29, 1506–1514 (2009).
Shin, H., Kim, D. & Helfman, D. M. Tropomyosin isoform Tpm2.1 regulates collective and amoeboid cell migration and cell aggregation in breast epithelial cells. Oncotarget 8, 95192–95205 (2017).
Lees, J. G. et al. Tropomyosin regulates cell migration during skin wound healing. J. Invest. Dermatol. 133, 1330–1339 (2013).
Koning, R. I., Koster, A. J. & Sharp, T. H. Advances in cryo-electron tomography for biology and medicine. Ann. Anat. 217, 82–96 (2018).
Simpson, L. J., Reader, J. S. & Tzima, E. Mechanical forces and their effect on the ribosome and protein translation machinery. Cells https://doi.org/10.3390/cells9030650 (2020).
Stahnke, S. et al. Loss of Hem1 disrupts macrophage function and impacts migration, phagocytosis, and integrin-mediated adhesion. Curr. Biol. https://doi.org/10.1016/j.cub.2021.02.043 (2021).
Kim, D. H. & Wirtz, D. Focal adhesion size uniquely predicts cell migration. FASEB J. 27, 1351–1361 (2013).
Meiring, J. C. M. et al. Co-polymers of actin and tropomyosin account for a major fraction of the human actin cytoskeleton. Curr. Biol. 28, 2331–2337.e2335 (2018).
Prasad, G. L., Fuldner, R. A. & Cooper, H. L. Expression of transduced tropomyosin 1 cDNA suppresses neoplastic growth of cells transformed by the ras oncogene. Proc. Natl Acad. Sci. USA 90, 7039–7043 (1993).
Cramer, L. P., Siebert, M. & Mitchison, T. J. Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 136, 1287–1305 (1997).
Naumanen, P., Lappalainen, P. & Hotulainen, P. Mechanisms of actin stress fibre assembly. J. Microsc. 231, 446–454 (2008).
Tojkander, S., Gateva, G. & Lappalainen, P. Actin stress fibers—assembly, dynamics and biological roles. J. Cell Sci. 125, 1855–1864 (2012).
Lazarides, E. & Burridge, K. α-Actinin: immunofluorescent localization of a muscle structural protein in nonmuscle cells. Cell 6, 289–298 (1975).
Weber, K. & Groeschel-Stewart, U. Antibody to myosin: the specific visualization of myosin-containing filaments in nonmuscle cells. Proc. Natl Acad. Sci. USA 71, 4561–4564 (1974).
Kemp, J. P. & Brieher, W. M. The actin filament bundling protein α-actinin-4 actually suppresses actin stress fibers by permitting actin turnover. J. Biol. Chem. 293, 14520–14533 (2018).
Meiring, J. C. M. et al. Colocation of Tpm3.1 and myosin IIa heads defines a discrete subdomain in stress fibres. J. Cell Sci. https://doi.org/10.1242/jcs.228916 (2019).
Elosegui-Artola, A. et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18, 540–548 (2016).
Pollard, T. D. Reflections on a quarter century of research on contractile systems. Trends Biochem. Sci. 25, 607–611 (2000).
Chitty, J. L. et al. The Mini‐Organo: a rapid high-throughput 3D coculture organotypic assay for oncology screening and drug development. Cancer Rep. https://doi.org/10.1002/cnr2.1209 (2020).
Bell, E., Ivarsson, B. & Merrill, C. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proc. Natl Acad. Sci. USA 76, 1274–1278 (1979).
Vogel, V. & Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275 (2006).
Iskratsch, T., Wolfenson, H. & Sheetz, M. P. Appreciating force and shape—the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 15, 825–833 (2014).
Hardeman, E. C. & Gunning, P. W. Life and death agendas of actin filaments. Nat. Mater. 19, 135–136 (2020).
Molinie, N. & Gautreau, A. The Arp2/3 regulatory system and its deregulation in cancer. Physiol. Rev. 98, 215–238 (2018).
Hendricks, M. & Weintraub, H. Tropomyosin is decreased in transformed cells. Proc. Natl Acad. Sci. USA 78, 5633–5637 (1981).
Coombes, J. D. et al. Ras transformation overrides a proliferation defect induced by Tpm3.1 knockout. Cell. Mol. Biol. Lett. 20, 626–646 (2015).
Hahn, W. C. et al. Creation of human tumour cells with defined genetic elements. Nature 400, 464–468 (1999).
Ponten, J. & Saksela, E. Two established in vitro cell lines from human mesenchymal tumours. Int. J. Cancer 2, 434–447 (1967).
Schevzov, G., Whittaker, S. P., Fath, T., Lin, J. J. & Gunning, P. W. Tropomyosin isoforms and reagents. Bioarchitecture 1, 135–164 (2011).
Acknowledgements
We would like to thank the Katharina Gaus Light Microscopy Facility, the Flow Cytometry Facility and the Electron Microscopy Unit of the Mark Wainwright Analytical Centre at UNSW, and the Cryo Electron Microscopy Facility through the Victor Chang Innovation Centre for their valuable help. Thanks to J. Hook and Y. Yao for their technical assistance. P.W.G. and E.C.H. were supported by grants from the ARC (DP160101623), the Australian NHMRC (APP1100202, APP1079866) and The Kid’s Cancer Project. N.A. was supported by the Australian NHMRC (APP1102730).
Author information
Authors and Affiliations
Contributions
M.L.C. conceptualized and wrote the manuscript. M.L.C., N.A. and S.B. performed the data acquisition and analysis. P.W.G., E.C.H., N.S.B. and N.A. reviewed the manuscript, supervised the work and acquired funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Materials thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7 and Video 1.
41563_2021_1087_MOESM3_ESM.mp4
Supplementary Video 1 Cryo-tomogram of a nascent adhesion site. Related to Fig. 2. Notice the increased cellular density at the filaments/adhesion Z-plane compared to the cytoplasm above
Source data
Source Data Fig. 4
Source Data including uncropped blots has been provided as a pdf via the uploading link
Rights and permissions
About this article
Cite this article
Cagigas, M.L., Bryce, N.S., Ariotti, N. et al. Correlative cryo-ET identifies actin/tropomyosin filaments that mediate cell–substrate adhesion in cancer cells and mechanosensitivity of cell proliferation. Nat. Mater. 21, 120–128 (2022). https://doi.org/10.1038/s41563-021-01087-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-021-01087-z
This article is cited by
-
Tropomyosin1 isoforms underlie epithelial to mesenchymal plasticity, metastatic dissemination, and resistance to chemotherapy in high-grade serous ovarian cancer
Cell Death & Differentiation (2024)
-
Focal adhesions contain three specialized actin nanoscale layers
Nature Communications (2024)
-
TPM2 attenuates progression of prostate cancer by blocking PDLIM7-mediated nuclear translocation of YAP1
Cell & Bioscience (2023)
-
Identification of a novel circRNA–miRNA–mRNA regulatory axis in hepatocellular carcinoma based on bioinformatics analysis
Scientific Reports (2023)
-
Impairment of rigidity sensing caused by mutant TP53 gain of function in osteosarcoma
Bone Research (2023)