Abstract
Actomyosin machinery endows cells with contractility at a single-cell level. However, within a monolayer, cells can be contractile or extensile based on the direction of pushing or pulling forces exerted by their neighbours or on the substrate. It has been shown that a monolayer of fibroblasts behaves as a contractile system while epithelial or neural progentior monolayers behave as an extensile system. Through a combination of cell culture experiments and in silico modelling, we reveal the mechanism behind this switch in extensile to contractile as the weakening of intercellular contacts. This switch promotes the build-up of tension at the cell–substrate interface through an increase in actin stress fibres and traction forces. This is accompanied by mechanotransductive changes in vinculin and YAP activation. We further show that contractile and extensile differences in cell activity sort cells in mixtures, uncovering a generic mechanism for pattern formation during cell competition, and morphogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data for cell orientation detection, and Bayesian inference of tissue stress are available upon request from the corresponding author. Source data are provided with this paper.
Code availability
Codes for cell orientation detection, and Bayesian inference of tissue stress are available upon request from the corresponding author. Numerical analyses were performed using a custom-made code: CELADRO (Cells as Active Droplets), which is open source and has been deposited on GitHub (https://github.com/rhomu/celadro).
Change history
09 March 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41563-021-00974-9
References
Barriga, E. H., Franze, K., Charras, G. & Mayor, R. Tissue stiffening coordinates morphogenesis by triggering collective cell migration in vivo. Nature 554, 523–527 (2018).
Onodera, T. et al. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science 329, 562–565 (2010).
Chen, X. L. et al. VEGF-induced vascular permeability is mediated by FAK. Dev. Cell 22, 146–157 (2012).
Malinverno, C. et al. Endocytic reawakening of motility in jammed epithelia. Nat. Mater. 16, 587–596 (2017).
Cetera, M. et al. Epithelial rotation promotes the global alignment of contractile actin bundles during Drosophila egg chamber elongation. Nat. Commun. 5, 5511 (2014).
Wang, S., Matsumoto, K. & Yamada, K. M. Reconstituting stratified epithelial branching morphogenesis by engineering cell adhesion. Preprint at bioRxiv https://doi.org/10.1101/2020.06.24.165795v1 (2020).
De Pascalis, C. & Etienne-Manneville, S. Single and collective cell migration: the mechanics of adhesions. Mol. Biol. Cell 28, 1833–1846 (2017).
Schwarz, U. S. & Safran, S. A. Elastic interactions of cells. Phys. Rev. Lett. 88, 048102 (2002).
Duclos, G., Erlenkämper, C., Joanny, J.-F. & Silberzan, P. Topological defects in confined populations of spindle-shaped cells. Nat. Phys. 13, 58–62 (2017).
Saw, T. B. et al. Topological defects in epithelia govern cell death and extrusion. Nature 544, 212–216 (2017).
Blanch-Mercader, C. et al. Turbulent dynamics of epithelial cell cultures. Phys. Rev. Lett. 120, 208101 (2018).
Kawaguchi, K., Kageyama, R. & Sano, M. Topological defects control collective dynamics in neural progenitor cell cultures. Nature 545, 327–331 (2017).
Xi, W., Saw, T. B., Delacour, D., Lim, C. T. & Ladoux, B. Material approaches to active tissue mechanics. Nat. Rev. Mater. 4, 23–44 (2019).
Gruler, H., Dewald, U. & Eberhardt, M. Nematic liquid crystals formed by living amoeboid cells. Eur. Phys. J. B 11, 187–192 (1999).
Ladoux, B. & Mège, R.-M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 18, 743–757 (2017).
Giomi, L., Bowick, M. J., Mishra, P., Sknepnek, R. & Cristina Marchetti, M. Defect dynamics in active nematics. Philos. Trans. R. Soc. A 372, 20130365 (2014).
Doostmohammadi, A., Ignés-Mullol, J., Yeomans, J. M. & Sagués, F. Active nematics. Nat. Commun. 9, 3246 (2018).
Marth, W. & Voigt, A. Signaling networks and cell motility: a computational approach using a phase field description. J. Math. Biol. 69, 91–112 (2014).
Mueller, R., Yeomans, J. M. & Doostmohammadi, A. Emergence of active nematic behavior in monolayers of isotropic cells. Phys. Rev. Lett. 122, 048004 (2019).
Ng, M. R., Besser, A., Brugge, J. S. & Danuser, G. Mapping the dynamics of force transduction at cell–cell junctions of epithelial clusters. eLife 3, e03282 (2014).
Nier, V. et al. Inference of internal stress in a cell monolayer. Biophys. J. 110, 1625–1635 (2016).
Petitjean, L. et al. Velocity fields in a collectively migrating epithelium. Biophys. J. 98, 1790–1800 (2010).
Garcia, S. et al. Physics of active jamming during collective cellular motion in a monolayer. Proc. Natl Acad. Sci. USA 112, 15314–15319 (2015).
Goodwin, K. et al. Cell–cell and cell–extracellular matrix adhesions cooperate to organize actomyosin networks and maintain force transmission during dorsal closure. Mol. Biol. Cell 28, 1301–1310 (2017).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Giannone, G. et al. Periodic lamellipodial contractions correlate with rearward actin waves. Cell 116, 431–443 (2004).
Zemel, A., Rehfeldt, F., Brown, A. E. X., Discher, D. E. & Safran, S. A. Optimal matrix rigidity for stress-fibre polarization in stem cells. Nat. Phys. 6, 468–473 (2010).
Gupta, M. et al. Adaptive rheology and ordering of cell cytoskeleton govern matrix rigidity sensing. Nat. Commun. 6, 7525 (2015).
Saraswathibhatla, A. & Notbohm, J. Tractions and stress fibers control cell shape and rearrangements in collective cell migration. Phys. Rev. X 10, 011016 (2020).
Riveline, D. et al. Focal contacts as mechanosensors. J. Cell Biol. 153, 1175–1186 (2001).
Yonemura, S., Wada, Y., Watanabe, T., Nagafuchi, A. & Shibata, M. α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 (2010).
Bays, J. L. & DeMali, K. A. Vinculin in cell–cell and cell–matrix adhesions. Cell. Mol. Life Sci. 74, 2999–3009 (2017).
Halder, G., Dupont, S. & Piccolo, S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 13, 591–600 (2012).
Benham-Pyle, B. W., Pruitt, B. L. & Nelson, W. J. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 348, 1024–1027 (2015).
Kim, N.-G., Koh, E., Chen, X. & Gumbiner, B. M. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl Acad. Sci. USA 108, 11930–11935 (2011).
Discher, D. E., Janmey, P. & Wang, Y.-l Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 (2005).
Maître, J.-L. et al. Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science 338, 253–256 (2012).
Steinberg, M. S. Does differential adhesion govern self-assembly processes in histogenesis? Equilibrium configurations and the emergence of a hierarchy among populations of embryonic cells. J. Exp. Zool. 173, 395–433 (1970).
Manning, M. L., Foty, R. A., Steinberg, M. S. & Schoetz, E.-M. Coaction of intercellular adhesion and cortical tension specifies tissue surface tension. Proc. Natl Acad. Sci. USA 107, 12517–12522 (2010).
Sahu, P. et al. Small-scale demixing in confluent biological tissues. Soft Matter 16, 3325–3337 (2020).
Krajnc, M. Solid-fluid transition and cell sorting in epithelia with junctional tension fluctuations. Soft Matter 16, 3209–3215 (2020).
Thomas, W. A., Thomson, J., Magnani, J. L. & Steinberg, M. S. Two distinct adhesion mechanisms in embryonic neural retina cells: III. Functional specificity. Dev. Biol. 81, 379–385 (1981).
Foty, R. A. & Steinberg, M. S. Cadherin-mediated cell-cell adhesion and tissue segregation in relation to malignancy. Int. J. Dev. Biol. 48, 397–409 (2004).
Niessen, C. M. & Gumbiner, B. M. Cadherin-mediated cell sorting not determined by binding or adhesion specificity. J. Cell Biol. 156, 389–400 (2002).
Maroudas-Sacks, Y. et al. Topological defects in the nematic order of actin fibers as organization centers of Hydra morphogenesis. Nat. Phys. https://doi.org/10.1038/s41567-020-01083-1 (2020).
Comelles, J. et al. Epithelial colonies in vitro elongate through collective effects. eLife 10, e57730 (2020).
Morales-Navarrete, H. et al. Liquid-crystal organization of liver tissue. eLife 8, e44860 (2019).
Aigouy, B. et al. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell 142, 773–786 (2010).
Hannezo, E., Prost, J. & Joanny, J.-F. Theory of epithelial sheet morphology in three dimensions. Proc. Natl Acad. Sci. USA 111, 27–32 (2014).
Doostmohammadi, A. et al. Celebrating soft matter’s 10th anniversary: cell division: a source of active stress in cellular monolayers. Soft Matter 11, 7328–7336 (2015).
Marthiens, V., Padilla, F., Lambert, M. & Mege, R. M. Complementary expression and regulation of cadherins 6 and 11 during specific steps of motoneuron differentiation. Mol. Cell. Neurosci. 20, 458–475 (2002).
Glukhova, M. A., Frid, M. G. & Koteliansky, V. E. Developmental changes in expression of contractile and cytoskeletal proteins in human aortic smooth muscle. J. Biol. Chem. 265, 13042–13046 (1990).
Vedula, S. R. K. et al. Epithelial bridges maintain tissue integrity during collective cell migration. Nat. Mater. 13, 87–96 (2014).
Hara, Y., Shagirov, M. & Toyama, Y. Cell boundary elongation by non-autonomous contractility in cell oscillation. Curr. Biol. 26, 2388–2396 (2016).
Meijering, E., Dzyubachyk, O. & Smal, I. Methods for cell and particle tracking. Meth. Enzymol. 504, 183–200 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Meth. 9, 676–682 (2012).
Plotnikov, S. V., Sabass, B., Schwarz, U. S. & Waterman, C. M. High-resolution traction force microscopy. Meth. Cell Biol. 123, 367–394 (2014).
Tseng, Q. et al. Spatial organization of the extracellular matrix regulates cell–cell junction positioning. Proc. Natl Acad. Sci. USA 109, 1506–1511 (2012).
Peyret, G. et al. Sustained oscillations of epithelial cell sheets. Biophys. J. 117, 464–478 (2019).
Aronson, I. S. (ed.) Physical Models of Cell Motility. Biological and Medical Physics, Biomedical Engineering (Springer, 2016).
Camley, B. A. & Rappel, W.-J. Physical models of collective cell motility: from cell to tissue. J. Phys. D 50, 113002 (2017).
Palmieri, B., Bresler, Y., Wirtz, D. & Grant, M. Multiple scale model for cell migration in monolayers: elastic mismatch between cells enhances motility. Sci. Rep. 5, 11745 (2015).
Winkler, B., Aranson, I. S. & Ziebert, F. Confinement and substrate topography control cell migration in a 3D computational model. Commun. Phys. 2, 82 (2019).
Alert, R. & Trepat, X. Physical models of collective cell migration. Annu. Rev. Condens. Matter Phys. 11, 77–101 (2020).
Smeets, B. et al. Emergent structures and dynamics of cell colonies by contact inhibition of locomotion. Proc. Natl Acad. Sci. USA 113, 14621–14626 (2016).
Acknowledgements
This work was supported by the European Research Council (grant no. CoG-617233), LABEX Who Am I? (ANR-11-LABX-0071), the Ligue Contre le Cancer (Equipe labellisée), and the Agence Nationale de la Recherche (‘POLCAM’ (ANR-17-CE13-0013 and ‘MechanoAdipo’ ANR-17-CE13-0012). We acknowledge the ImagoSeine core facility of the IJM, member of IBiSA and France-BioImaging (ANR-10-INBS-04) infrastructures. A.D. acknowledges support from the Novo Nordisk Foundation (grant no. NNF18SA0035142), Villum Fonden (grant no. 29476), and funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement no. 847523 (INTERACTIONS). L.B. has received funding from the European Union’s Horizon 2020 research and innovation programme (Marie Sklodowska-Curie grant agreement no. 665850-INSPIRE). T.B.S. acknowledges support from a Lee Kuan Yew (LKY) postdoctoral fellowship and the Singapore Ministry of Education Academic Research Fund Tier 1 (R-397-000-320-114). S.G. and A.Y. were supported by project grants and fellowships from the National Health and Medical Research Council of Australia (1123816 and 1139592) and Australian Research Council (DP190102871). We would like to thank P. Marcq for help with implementation of BISM code, M. A. Glukhova for providing the vinculin antibody and S. Robine for the ZO1 antibody. A.D. and J.M.Y. acknowledge G. Zhang for helpful discussions regarding the model. We also thank the members of cell adhesion and mechanics team at Institut Jacques Monod, M. Piel and F. Gallet, for insightful discussions.
Author information
Authors and Affiliations
Contributions
L.B., T.B.S., A.D., J.Y., R.M.M. and B.L. designed the research. G.H.N.S.N. and T.D. developed the E-cadherin KO cell line. L.B. performed experiments and analysed the results. S.S. helped in the PA gel experiments. M.T. performed and quantified laser ablation experiments. S.G. performed the MCF7 experiments. A.D. and R.M. performed the in silico simulations. T.B.S. contributed to the analysis tools. A.S.Y., Y.T., R.M.M., A.D., J.Y. and B.L supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Materials thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 MDCK WT behave as an extensile system.
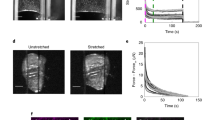
a, Kymograph of a short junction (<10μm) (top) and long junction (>15μm) (bottom) before and after laser ablation. b, Recoil velocity after laser ablation for short (<10μm) (n=9) (N=4), medium (10-15μm) (n=13)(N=4) and long junctions (> 15μm) (n=12) (N=6). n, is the number of junctions ablated and N is the number of independent experiments from which these results were obtained. Error bars represent the standard deviation. ANOVA test was performed leading to *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001. Scale bars, 20μm.
Extended Data Fig. 2 MDCK WT behaves as an extensile system and MDCK E-cadherin KO behave as a contractile system.
a,b, Orientation field (left) and velocity vectors (right) around a single comet-shaped (+1/2) defect (a) and trefoil (-1/2) defect obtained from WT (top) and E-cadherin KO (bottom) monolayers. c,d, Trajectory of several comet (+1/2) (left) and trefoil (-1/2) (right) shaped defects obtained from MDCK WT (c) and MDCK E-cadherin KO (d) monolayers. Scale bars: 40μm.
Extended Data Fig. 3 Fibroblasts behave as a 2D contractile active nematic.
a, Average yy- and xy-components of strain rate map around +1/2 defect obtained from experiments (left and middle respectively) and corresponding average flow field (right) (n = 1489 defects from 2 independent experiments) for NIH3T3 cells. Colour code is positive for stretching and negative for shrinkage. b, Average yy (left)-, xy (middle)- and isotropic (right) components of stress around a +1/2 defect obtained from experiments for NIH3T3 (n = 1,428 defects from 2 independent experiments).
Extended Data Fig. 4 Characterization of MDCK E-cadherin KO cells and extensile/contractile behaviour of MCF7 cells.
a, Immunofluorescence staining (top) of E-cadherin (left), β-catenin (middle) and ZO1 (right), along with a b) representative western blot and quantification for E-cadherin (left) (n=3) and β-catenin (right) (n=3). Scale bars, 20μm. c, Western blot analysis of total MLC and quantification from 3 independent experiments normalized to α-tubulin. d, Western blot analysis of vinculin and quantification from 3 independent experiments normalized to GAPDH. Error bars represent the standard deviation. e, Western blot showing the reduced level of E-cadherin in siRNA generated E-cadherin KD cell line for MCF7 cells. f,g Average flow field for MCF7 control cells (n = 2047 defects from 3 independent experiments) (f) and siRNA E-cadherin KD MCF7 cells (n = 1256 defects from 3 independent experiments) (f). h, Uncropped blots of all the western blots shown so far.
Extended Data Fig. 5 E-cadherin rescue changes the behaviour to a 2D extensile active nematic liquid crystal and flow field around trefoil shaped defects.
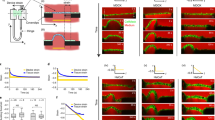
a, Average yy- and xy-components of strain rate map around comet (+1/2) defect obtained from experiments (left and middle respectively) and corresponding average flow field (right) (n = 1767 defects from 2 independent experiments) for MDCK E-cadherin KO cells rescued with E-cadherin GFP. b, Total number of defects obtained per 0.55mm2 as a function of time on MDCK WT and MDCK E-cadherin KO monolayers. (n=10) from 2 independent experiments. c, Average vorticity and velocity field around trefoil (-1/2) defects in WT (left) (n=1934) and E-cadherin KO (right) (n=2028) monolayers. d, Average vorticity and velocity field around trefoil (-1/2) defects in control (left) (n=3200) and condition without active intercellular forces (right)(n=3200) monolayers obtained from simulations. e, Mean square displacement (MSD) plotted against time lag for comet (+1/2) and trefoil (-1/2) defects obtained from MDCK WT and MDCK E-cadherin KO monolayers (n=11). Error bars represent the standard deviation.
Extended Data Fig. 6 E-cadherin removal does not affect the contractile behaviour of single cells.
a, Mean traction force for both MDCK WT (n=31) and MDCK E-cadherin KO cells (n=27). b, Average yy- and xy-components of strain rate map around comet (+1/2) defect obtained from experiments (left and middle respectively) and corresponding average velocity flow field (right) (n = 1428 defects from 2 independent experiments) for MDCK E-cadherin KO cells plated on PDMS substrates of stiffness 15kPa from which stress maps were obtained in Fig. 3b’.
Extended Data Fig. 7 Drug treatment changes 2D active nematic behaviour of MDCK E-cadherin KO cells.
a,b,c, Average yy- and xy-components of strain rate map around +1/2 defect obtained from experiments (left and middle respectively) and corresponding average velocity flow field (right) for MDCK E-cadherin KO cells treated with 5μM blebbistatin (a) (n = 2174 defects from 2 independent experiments), 20μM blebbistatin (b) (n = 1223 defects from 2 independent experiments), and 25μM Y27632 (c) (n = 1965 defects from 2 independent experiments). d,e, Average yy- and xy components of strain rate map around +1/2 defect obtained from experiments (left and middle respectively) and corresponding average velocity flow field (right) for MDCK WT cells treated with 20μM blebbistatin (d) (n = 1287 defects from 2 independent experiments), and 25μM Y27632 (e) (n = 2472 defects from 2 independent experiments).
Extended Data Fig. 8 Colocalization of vinculin and paxillin.
a, Immunostaining of basal plane of vinculin (left), paxillin (middle) and merge (right) in MDCK WT (top) and MDCK E-cadherin KO monolayers. b, Intensity of vinculin plotted against paxillin for n=15 focal adhesions in MDCK WT and n=16 focal adhesions in MDCK E-cadherin KO monolayers. Scale bars: 20μm.
Extended Data Fig. 9 Substrate rigidity alters E-cadherin KO behaviour.
a,b, Average yy- and xy-components of strain rate map around +1/2 defect obtained from experiments (left and middle respectively) and corresponding average flow field (right) for MDCK WT cells (a) (n = 1426 defects from 2 independent experiments) and E-cadherin KO cells (b) (n = 1041 defects from 2 independent experiments).
Extended Data Fig. 10 Phase diagram on activity change and activity based cell sorting in a mixed culture of MDCK WT and MDCK E-cadherin KO.
a, Phase diagram showing the transition of extensile and contractile behaviour with varying values of intercellular and intracellular stresses obtained from simulations. b, Phase separation (demixing) observed from simulations where the contractile particles (orange) are surrounded by extensile particles (green). c, Cell sorting (demixing) observed for a mixture of MDCK WT and MDCK E-cadherin KO cells, where WT cells are surrounded by E-cadherin KO cells (E-cadherin, green, cadherin 6, red, actin, black). XZ and YZ projection show the height difference between the two cells when mixed. Scale bars, 20μm. d,e, Early stages of cell sorting when MDCK WT (magenta) and MDCK E-cadherin KO (green) monolayers are mixed at 30-70 (d) and 70-30 (e) ratio. Scale bars: 100μm.
Supplementary information
Supplementary Information
Supplementary Table 1 and video legends
Supplementary Video 1
Phase contrast images overlaid with orientation vectors obtained from MDCK WT monolayers. Blue arrow shows the location of a +1/2 (comet)-shaped defect and the direction in which they move. Scale bar: 50 μm.
Supplementary Video 2
Phase contrast images overlaid with orientation vectors obtained from MDCK E-cadherin KO monolayers. Blue arrow shows the location of a +1/2 (comet)-shaped defect and the direction in which they move. Scale bar: 50 μm.
Supplementary Video 3
Time-lapse demixing of extensile (magenta) and contractile (green) particles obtained from simulations.
Supplementary Video 4
Time-lapse demixing of MDCK WT (extensile-magenta) and MDCK E-cadherin KO (contractile-green) cells observed from experiments. Scale bar: 100 μm.
Supplementary Video 5
Time-lapse demixing of MDCK WT (extensile-magenta) and MDCK E-cadherin KO (contractile-green) cells before and after the addition of 20 μM blebbistatin. Scale bar: 100 μm.
Source data
Source Data Fig. 2
Data used to plot graphs.
Source Data Fig. 3
Data used to plot graphs.
Source Data Fig. 4
Data used to plot graphs.
Source Data Fig. 5
Data used to plot graphs.
Source Data Fig. 6
Data used to plot graphs.
Source Data Extended Data Fig. 1
Data used to plot graphs.
Source Data Extended Data Fig. 4
Data used to plot graphs and uncropped western blots.
Source Data Extended Data Fig. 5
Data used to plot graphs.
Source Data Extended Data Fig. 6
Data used to plot graphs.
Source Data Extended Data Fig. 9
Data used to plot graphs.
Rights and permissions
About this article
Cite this article
Balasubramaniam, L., Doostmohammadi, A., Saw, T.B. et al. Investigating the nature of active forces in tissues reveals how contractile cells can form extensile monolayers. Nat. Mater. 20, 1156–1166 (2021). https://doi.org/10.1038/s41563-021-00919-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-021-00919-2
This article is cited by
-
Photosynthetically-powered phototactic active nematic liquid crystal fluids and gels
Communications Materials (2024)
-
How multiscale curvature couples forces to cellular functions
Nature Reviews Physics (2024)
-
Defect-mediated dynamics of coherent structures in active nematics
Nature Physics (2023)
-
Mechanical stress driven by rigidity sensing governs epithelial stability
Nature Physics (2023)
-
Hexanematic crossover in epithelial monolayers depends on cell adhesion and cell density
Nature Communications (2023)