Abstract
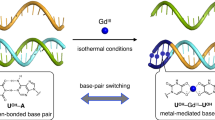
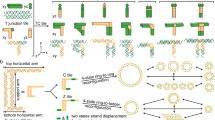
The diversity of DNA duplex structures is limited by a binary pair of hydrogen-bonded motifs. Here we show that poly(thymine) self-associates into antiparallel, right-handed duplexes in the presence of melamine, a small molecule that presents a triplicate set of the hydrogen-bonding face of adenine. X-ray crystallography shows that in the complex two poly(thymine) strands wrap around a helical column of melamine, which hydrogen bonds to thymine residues on two of its three faces. The mechanical strength of the thymine–melamine–thymine triplet surpasses that of adenine–thymine base pairs, which enables a sensitive detection of melamine at 3 pM. The poly(thymine)–melamine duplex is orthogonal to native DNA base pairing and can undergo strand displacement without the need for overhangs. Its incorporation into two-dimensional grids and hybrid DNA–small-molecule polymers highlights the poly(thymine)–melamine duplex as an additional tool for DNA nanotechnology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Crystallography data (Fig. 3) are available from the Protein Data Bank (https://www.rcsb.org/) with access code 6WK7. The source data related to Figs. 4 and 5 are available upon request from the corresponding authors. All other data in the paper are provided as Source data.
References
Frank-Kamenetskii, M. D. & Mirkin, S. M. Triplex DNA structures. Annu. Rev. Biochem. 64, 65–95 (1995).
Rhodes, D. & Lipps, H. J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 43, 8627–8637 (2015).
Abou Assi, H., Garavís, M., González, C. & Damha, M. J. i-motif DNA: structural features and significance to cell biology. Nucleic Acids Res. 46, 8038–8056 (2018).
Debnath, M., Fatma, K. & Dash, J. Chemical regulation of DNA i-motifs for nanobiotechnology and therapeutics. Angew. Chem. Int. Ed. 58, 2942–2957 (2019).
Kang, S., Ohshima, K., Shimizu, M., Amirhaeri, S. & Wells, R. D. Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem. 270, 27014–27021 (1995).
Dong, Y., Yang, Z. & Liu, D. Using small molecules to prepare vesicles with designable shapes and sizes via frame-guided assembly strategy. Small 11, 3768–3771 (2015).
Davies, R. J. H. & Davidson, N. Base pairing equilibria between polynucleotides and complementary monomers. Biopolymers 10, 1455–1479 (1971).
Li, C., Cafferty, B. J., Karunakaran, S. C., Schuster, G. B. & Hud, N. V. Formation of supramolecular assemblies and liquid crystals by purine nucleobases and cyanuric acid in water: implications for the possible origins of RNA. Phys. Chem. Chem. Phys. 18, 20091–20096 (2016).
Chen, D., Meena, Sharma, S. K. & McLaughlin, L. W. Formation and stability of a Janus-wedge type of DNA triplex. J. Am. Chem. Soc. 126, 70–71 (2004).
Zeng, Y., Pratumyot, Y., Piao, X. & Bong, D. Discrete assembly of synthetic peptide–DNA triplex structures from polyvalent melamine–thymine bifacial recognition. J. Am. Chem. Soc. 134, 832–835 (2012).
Avakyan, N. et al. Reprogramming the assembly of unmodified DNA with a small molecule. Nat. Chem. 8, 368–376 (2016).
Zhou, W., Saran, R. & Liu, J. Metal sensing by DNA. Chem. Rev. 117, 8272–8325 (2017).
Gothelf, K. V., Thomsen, A., Nielsen, M., Cló, E. & Brown, R. S. Modular DNA-programmed assembly of linear and branched conjugated nanostructures. J. Am. Chem. Soc. 126, 1044–1046 (2004).
Yamane, T. & Davidson, N. On the complexing of desoxyribonucleic acid (DNA) by mercuric ion. J. Am. Chem. Soc. 83, 2599–2607 (1961).
Ono, A. et al. Specific interactions between silver(i) ions and cytosine–cytosine pairs in DNA duplexes. Chem. Commun. 2008, 4825–4827 (2008).
Tanaka, K., Tengeiji, A., Kato, T., Toyama, N. & Shionoya, M. A discrete self-assembled metal array in artificial DNA. Science 299, 1212–1213 (2003).
Mao, J., DeSantis, C. & Bong, D. Small molecule recognition triggers secondary and tertiary interactions in DNA folding and hammerhead ribozyme catalysis. J. Am. Chem. Soc. 139, 9815–9818 (2017).
Vollhardt, D., Liu, F., Rudert, R. & He, W. Interfacial molecular recognition of dissolved thymine by medium chain dialkyl melamine-type monolayers. J. Phys. Chem. B 109, 10849–10857 (2005).
Lange, R. F. M. et al. Crystal engineering of melamine–Imide complexes; tuning the stoichiometry by steric hindrance of the imide carbonyl groups. Angew. Chem. Int. Ed. 36, 969–971 (1997).
Du, J., Wang, Z., Peng, X. & Fan, J. In situ colorimetric recognition of melamine based on thymine derivative-functionalized gold nanoparticle. Ind. Eng. Chem. Res. 54, 12011–12016 (2015).
Renny, J. S., Tomasevich, L. L., Tallmadge, E. H. & Collum, D. B. Method of continuous variations: applications of job plots to the study of molecular associations in organometallic chemistry. Angew. Chem. Int. Ed. 52, 11998–12013 (2013).
Ulatowski, F., Dąbrowa, K., Bałakier, T. & Jurczak, J. Recognizing the limited applicability of job plots in studying host–guest interactions in supramolecular chemistry. J. Org. Chem. 81, 1746–1756 (2016).
Mikol, V., Rodeau, J.-L. & Giegé, R. Experimental determination of water equilibration rates in the hanging drop method of protein crystallization. Anal. Biochem. 186, 332–339 (1990).
Dickerson, R. E. DNA structure from A to Z. Methods Enzymol. 211, 67–111 (1992).
Dixon, J. K., Woodberry, N. T. & Costa, G. W. The dissociation constants of melamine and certain of its compounds. J. Am. Chem. Soc. 69, 599–603 (1947).
De Greef, T. F. A. et al. Supramolecular polymerization. Chem. Rev. 109, 5687–5754 (2009).
Smulders, M. M. J. et al. How to distinguish isodesmic from cooperative supramolecular polymerisation. Chem. Eur. J. 16, 362–367 (2010).
Zhao, D. & Moore, J. S. Nucleation–elongation: a mechanism for cooperative supramolecular polymerization. Org. Biomol. Chem. 1, 3471–3491 (2003).
Seto, C. T. & Whitesides, G. M. Self-assembly based on the cyanuric acid–melamine lattice. J. Am. Chem. Soc. 112, 6409–6411 (1990).
Woodside, M. T. et al. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl Acad. Sci. USA 103, 6190–6195 (2006).
Koirala, D. et al. A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nat. Chem. 3, 782–787 (2011).
Jonchhe, S. et al. Binding of a telomestatin derivative changes mechanical anisotropy of human telomeric G-quadruplex. Angew. Chem. Int. Ed. Engl. 58, 877–881 (2019).
Koirala, D., Yu, Z., Dhakal, S. & Mao, H. Detection of single nucleotide polymorphism using tension-dependent stochastic behavior of a single-molecule template. J. Am. Chem. Soc. 133, 9988–9991 (2011).
Fire, A. & Xu, S. Q. Rolling replication of short DNA circles. Proc. Natl Acad. Sci. USA 92, 4641–4645 (1995).
Liu, D., Daubendiek, S. L., Zillman, M. A., Ryan, K. & Kool, E. T. Rolling circle DNA synthesis: small circular oligonucleotides as efficient templates for DNA polymerases. J. Am. Chem. Soc. 118, 1587–1594 (1996).
Mandal, S., Koirala, D., Selvam, S., Ghimire, C. & Mao, H. A molecular tuning fork in single-molecule mechanochemical sensing. Angew. Chem. Int. Ed. Engl. 54, 7607–7611 (2015).
Huang, H. et al. Visual detection of melamine in milk samples based on label-free and labeled gold nanoparticles. Talanta 85, 1013–1019 (2011).
Qi, W. J., Wu, D., Ling, J. & Huang, C. Z. Visual and light scattering spectrometric detections of melamine with polythymine-stabilized gold nanoparticles through specific triple hydrogen-bonding recognition. Chem. Commun. 46, 4893–4895 (2010).
Li, H., Somerson, J., Xia, F. & Plaxco, K. W. Electrochemical DNA-based sensors for molecular quality control: continuous, real-time melamine detection in flowing whole milk. Anal. Chem. 90, 10641–10645 (2018).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2017).
Fu, T. J. & Seeman, N. C. DNA double-crossover molecules. Biochemistry 32, 3211–3220 (1993).
Liu, L., Li, Y., Wang, Y., Zheng, J. & Mao, C. Regulating DNA self-assembly by DNA–surface interactions. ChemBioChem 18, 2404–2407 (2017).
Avakyan, N., Conway, J. W. & Sleiman, H. F. Long-range ordering of blunt-ended DNA tiles on supported lipid bilayers. J. Am. Chem. Soc. 139, 12027–12034 (2017).
He, Y., Chen, Y., Liu, H., Ribbe, A. E. & Mao, C. Self-assembly of hexagonal DNA two-dimensional (2D) arrays. J. Am. Chem. Soc. 127, 12202–12203 (2005).
He, Y. et al. Sequence symmetry as a tool for designing DNA nanostructures. Angew. Chem. Int. Ed. 44, 6694–6696 (2005).
Vantomme, G. & Meijer, E. W. The construction of supramolecular systems. Science 363, 1396–1397 (2019).
Sassolas, A., Leca-Bouvier, B. D. & Blum, L. J. DNA biosensors and microarrays. Chem. Rev. 108, 109–139 (2008).
Krishnamurthy, R. Experimentally investigating the origin of DNA/RNA on early Earth. Nat. Commun. 9, 5175 (2018).
Tarköy, M., Phipps, A. K., Schultze, P. & Feigon, J. Solution structure of an intramolecular DNA triplex linked by hexakis(ethylene glycol) units: d(AGAGAGAA-(EG)6-TTCTCTCT-(EG)6-TCTCTCTT). Biochemistry 37, 5810–5819 (1998).
Mao, H. & Luchette, P. An integrated laser-tweezers instrument for microanalysis of individual protein aggregates. Sens. Actuators B 129, 764–771 (2008).
Acknowledgements
This work was financial support by ONR (N00014-15-1-2707) and NSFC (21974111) to C.M., National Science Foundation (CBET-1904921) and National Institutes of Health (NIH 1R01CA236350) (in part) to H.M., the Natural Sciences and Engineering Research Council (NSERC) of Canada and the Canada Research Chairs Program to H.F.S. and F.J.R., and the Government of Canada for a Banting Fellowship to F.J.R.
Author information
Authors and Affiliations
Contributions
H.F.S., H.M. and C.M supervised the project. Q.L. and S.W. conducted PAGE analyses. J.Z. and H.H. conducted crystallographic analysis. L.L. conducted AFM imaging. S.J. and S.M. conducted mechanical measurements. Q.L. and F.J.R. conducted CD, thermal denaturation and assembly mechanism experiments. All the authors contributed to the data analysis and the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–26 and Tables 1–3.
Source data
Source Data Fig. 3
Source data for crystallography.
Source Data Fig. 4
Source data for mechanical force spectroscopy.
Source Data Fig. 5
Source data for mechanical force spectroscopy.
Rights and permissions
About this article
Cite this article
Li, Q., Zhao, J., Liu, L. et al. A poly(thymine)–melamine duplex for the assembly of DNA nanomaterials. Nat. Mater. 19, 1012–1018 (2020). https://doi.org/10.1038/s41563-020-0728-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-020-0728-2
This article is cited by
-
A dissipative pathway for the structural evolution of DNA fibres
Nature Chemistry (2021)