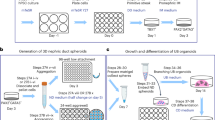
Abstract
Directed differentiation of human pluripotent stem cells to kidney organoids brings the prospect of drug screening, disease modelling and the generation of tissue for renal replacement. Currently, these applications are hampered by organoid variability, nephron immaturity, low throughput and limited scale. Here, we apply extrusion-based three-dimensional cellular bioprinting to deliver rapid and high-throughput generation of kidney organoids with highly reproducible cell number and viability. We demonstrate that manual organoid generation can be replaced by 6- or 96-well organoid bioprinting and evaluate the relative toxicity of aminoglycosides as a proof of concept for drug testing. In addition, three-dimensional bioprinting enables precise manipulation of biophysical properties, including organoid size, cell number and conformation, with modification of organoid conformation substantially increasing nephron yield per starting cell number. This facilitates the manufacture of uniformly patterned kidney tissue sheets with functional proximal tubular segments. Hence, automated extrusion-based bioprinting for kidney organoid production delivers improvements in throughput, quality control, scale and structure, facilitating in vitro and in vivo applications of stem cell-derived human kidney tissue.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All transcriptional profiling datasets have been submitted to GEO. These include scRNAseq from manual and two bioprinted organoid conformations (GEO GSE152014) and bulk-RNAseq data comparing bioprinted organoids of different conformations (GEO GSE138733). The image data used for quantification of morphology in different bioprinted organoid confirmations is available at https://doi.org/10.6084/m9.figshare.12957122.v1.
Code availability
Image and single cell RNAseq analysis scripts are available at http://github.com/KidneyRegeneration/BioprintedOrganoids/
References
Freedman, B. S. et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6, 8715 (2015).
Morizane, R. et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015).
Taguchi, A. et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14, 53–67 (2014).
Taguchi, A. & Nishinakamura, R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell 21, 730–746 (2017).
Little, M. H. & Combes, A. N. Kidney organoids: accurate models or fortunate accidents. Genes Dev. 33, 1319–1345 (2019).
Takasato, M., Er, P. X., Chiu, H. S. & Little, M. H. Generation of kidney organoids from human pluripotent stem cells. Nat. Protoc. 11, 1681–1692 (2016).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Jakab, K. et al. Tissue engineering by self-assembly and bio-printing of living cells. Biofabrication 2, 022001 (2010).
King, S. M. et al. 3D proximal tubule tissues recapitulate key aspects of renal physiology to enable nephrotoxicity testing. Front Physiol. 8, 123 (2017).
Nguyen, D. G. et al. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS ONE 11, e0158674 (2016).
Norona, L. M., Nguyen, D. G., Gerber, D. A., Presnell, S. C. & LeCluyse, E. L. Editor’s highlight: modeling compound-induced fibrogenesis in vitro using three-dimensional bioprinted human liver tissues. Toxicol. Sci. 154, 354–367 (2016).
Howden, S. E., Thomson, J. A. & Little, M. H. Simultaneous reprogramming and gene editing of human fibroblasts. Nat. Protoc. 13, 875–898 (2018).
Phipson, B. et al. Evaluation of variability in human kidney organoids. Nat. Methods 16, 79–87 (2019).
Hale, L. J. et al. 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat. Commun. 9, 5167 (2018).
Vanslambrouck, J. M. et al. A toolbox to characterize human induced pluripotent stem cell-derived kidney cell types and organoids. J. Am. Soc. Nephrol. 30, 1811–1823 (2019).
Soo, J. Y., Jansen, J., Masereeuw, R. & Little, M. H. Advances in predictive in vitro models of drug-induced nephrotoxicity. Nat. Rev. Nephrol. 14, 378–393 (2018).
Lin, Z. & Will, Y. Evaluation of drugs with specific organ toxicities in organ-specific cell lines. Toxicol. Sci. 126, 114–127 (2012).
Hallman, M. A., Zhuang, S. & Schnellmann, R. G. Regulation of dedifferentiation and redifferentiation in renal proximal tubular cells by the epidermal growth factor receptor. J. Pharmcol. Exp. Ther. 325, 520–528 (2008).
Vesey, D. A., Qi, W., Chen, X., Pollock, C. A. & Johnson, D. W. Isolation and primary culture of human proximal tubule cells. Methods Mol. Biol. 466, 19–24 (2009).
Wieser, M. et al. hTERT alone immortalizes epithelial cells of renal proximal tubules without changing their functional characteristics. Am. J. Physiol. Renal Physiol. 295, F1365–F1375 (2008).
Lee, V. W. & Harris, D. C. Adriamycin nephropathy: a model of focal segmental glomerulosclerosis. Nephrology (Carlton) 16, 30–38 (2011).
Su, Z., Ye, J., Qin, Z. & Ding, X. Protective effects of madecassoside against Doxorubicin induced nephrotoxicity in vivo and in vitro. Sci. Rep. 5, 18314 (2015).
Wargo, K. A. & Edwards, J. D. Aminoglycoside-induced nephrotoxicity. J. Pharm. Pract. 27, 573–577 (2014).
Ballatori, N. et al. OSTα-OSTβ: a major basolateral bile acid and steroid transporter in human intestinal, renal, and biliary epithelia. Hepatology 42, 1270–1279 (2005).
Lash, L. H., Putt, D. A. & Cai, H. Drug metabolism enzyme expression and activity in primary cultures of human proximal tubular cells. Toxicology 244, 56–65 (2008).
Maatman, R. G., Van Kuppevelt, T. H. & Veerkamp, J. H. Two types of fatty acid-binding protein in human kidney. Isolation, characterization and localization. Biochem J. 273, 759–766 (1991).
Palmiter, R. D. & Findley, S. D. Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 14, 639–649 (1995).
Sharmin, S. et al. Human induced pluripotent stem cell-derived podocytes mature into vascularized glomeruli upon experimental transplantation. J. Am. Soc. Nephrol. 27, 1778–1791 (2016).
van den Berg, C. W., Koudijs, A., Ritsma, L. & Rabelink, T. J. In vivo assessment of size-selective glomerular sieving in transplanted human induced pluripotent stem cell-derived kidney organoids. J. Am. Soc. Nephrol. 31, 921–929 (2020).
van den Berg, C. W. et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. 10, 751–765 (2018).
Combes, A. N., Zappia, L., Er, P. X., Oshlack, A. & Little, M. H. Single-cell analysis reveals congruence between kidney organoids and human fetal kidney. Genome Med. 11, 3 (2019).
Wu, H. et al. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell 23, 869–881 (2018).
Howden, S. E., Vanslambrouck, J. M., Wilson, S. B., Tan, K. S. & Little, M. H. Reporter-based fate mapping in human kidney organoids confirms nephron lineage relationships and reveals synchronous nephron formation. EMBO Rep. 20, e47483 (2019).
Hughson, M., Farris, A. B. III, Douglas-Denton, R., Hoy, W. E. & Bertram, J. F. Glomerular number and size in autopsy kidneys: the relationship to birth weight. Kidney Int. 63, 2113–2122 (2003).
Merlet-Benichou, C. et al. Nephron number: variability is the rule. Causes and consequences. Lab. Invest. 79, 515–527 (1999).
Czerniecki, S. M. et al. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 22, 929–940 (2018).
Chen, G. et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8, 424–429 (2011).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
van der Walt, S. et al. scikit-image: image processing in python. PeerJ 2, e453 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Acknowledgements
We thank M. Le Moing and the Murdoch Children’s Research Institute Translational Genomics Unit for 10x single cell and hash-tag oligo library preparation and sequencing and bulk-RNAseq. We thank M. Burton and the Murdoch Children’s Research Institute Microscopy Core for imaging support. M.H.L. is a Senior Principal Research Fellow of the National Health and Medical Research Council, Australia (APP1136085). This work was supported by Organovo Inc., the Methuselah Foundation, California’s Stem Cell Agency (EDUC2-08388), NHMRC (GNT1100970, GNT1098654), the National Institutes of Health (UH3DK107344) and a Medical Research Future Fund Kidney Disease Team grant.
Author information
Authors and Affiliations
Contributions
K.L., J.M.V., J.W.H., B.S., S.P., S.C.P., A.E.C. and M.H.L. contributed to the experimental design and planning. K.L., J.M.V., J.W.H., A.C., K.B., D.A., P.X.E., S.W., S.H., K.S.T., F.L. and L.J.H. developed methods and reagents, and performed and analysed experiments. All authors contributed to the interpretation of data. K.L., J.M.V., J.W.H., A.E.C. and M.H.L. contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.H.L. is an inventor on a patent related to kidney organoid generation. Access to the bioprinter was facilitated by the Methuselah Foundation. M.H.L. received contract research funding from Organovo Inc.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Histology of bioprinted kidney organoids.
a. Histological cross section of an entire day 7 + 18 bioprinted kidney organoid showing clear evidence of an interconnecting epithelium (arrowheads) from which nephrons arise. b. Immunostaining of a bioprinted kidney organoid section showing a GATA3 + ECAD + connecting segment / collecting duct with multiple attached ECAD + GATA3- nephrons. c. Immunostaining of bioprinted kidney organoid section showing ECAD + nephrons attached to MAFB + glomeruli. d. Brightfield, histological and immunofluorescence comparisons of kidney organoids generated manually (5 × 105 cells per organoid), using dry cell paste controlled for organoid diameter versus dry cell paste controlled for cell number versus wet cell paste. All image panels are representative of at least n = 3 organoids from multiple experiments.
Extended Data Fig. 2 Patterning of kidney structures in bioprinted organoids generated from varying starting cell numbers.
a. Immunofluorescence of organoids from a single starting differentiation used to generate manual organoids (5 × 105 cells) versus bioprinted organoids generated from as few as 4,000 cells. Representative images from n = 3 organoids stained. Scale bars represent 200 µm. b. Differentiation timecourse of bioprinted organoids generated using the MAFBmTagBFP2 reporter iPSC line. c. MAFBmTagBFP2 bioprinted organoids on the same Transwell filter with 4 K, 50 K or 100 K of cells per organoid showing fluorescence reporter imaging (blue) and staining for differentiation (ECAD,green; LTL, blue; GATA3, red; NPHS1, purple). Images are representative of at least n = 3 organoids. Scale bars represent 200 µm. d. MAFBmTagBFP2 bioprinted organoids on the same Transwell filter all generated using 100 K of cells per organoid showing live fluorescence imaging (blue) and staining for differentiation (ECAD,green; LTL, blue; GATA3, red; NPHS1, purple). Scale bars represent 200 µm. Representative wells from at least n = 3 are shown, with representative stained organoids alongside.
Extended Data Fig. 3 Quantification of bead density and MAFBmTagBFP2 reporter signal in organoids with varied conformations.
a. Representative image of fluorescent bead signal (greyscale) at D7 + 0 across an entire print pattern showing all 5 conformations, from left to right: ratio 0 (3 replicates), ratio 40, ratio 30, ratio 20, ratio 10. b. Composite image of each conformation at D7 + 12 showing mTagBFP2 reporter expression (cyan) and bead signal (red). Note images are placed on a black background. Scale bar is 1 mm for A and B. c. Quantification of total organoid area (refer to Methods) and mTagBFP2 area in replicate organoids (compare to Fig. 3g). d. Table of organoid numbers by replicate plate and ratio used for quantification in C and Fig. 3g. e. Example of 9 replicate organoids produced using ratio 20. Organoids are consistent between 3 organoids from separate wells on each plate, and between plates. f. Representative images (from total n = 27 organoids from 2 independent experiments) of sparse labelling with CellTrace Far Red dye to quantify organoid height at D7 + 0 (Fig. 3d). XY and orthogonal view are shown. g. Schematic of the scoring method used for quantification, described fully in Supplementary Methods.
Extended Data Fig. 4 MAFBmTagBFP2 reporter expression in organoids correlates to total nephron number.
a,b. Examples of low resolution, high throughput imaging used to quantify MAFB+ area as a proxy for nephron volume in organoids. Brightfield and MAFBmTagBFP2 signal was captured for each organoid using a low NA 4x objective with a spinning disk system, enabling fast capture of many samples. With a large axial depth of field, these images capture the majority of signal within each organoid in a single plane. Given the similarity in thickness (E,F, Fig. 3) this planar area is approximately proportional to total MAFB + glomerular volume and hence correlates to nephron number. A portion of an example image used for quantification of R0 (a) and R40 (b) organoids at D7 + 12 is shown. Note R40 organoids are much longer and were captured by stitching multiple image fields. Only a small portion of the organoid is shown. c,d. Samples were fixed and stained at D7 + 12 for MAFBmTagBFP2 reporter (Cyan), mature podocyte marker NPHS1 (Red) and atypical protein kinase C (aPKC, Green), a marker of the apical cell membrane. Each nephron consists of a rounded glomerular structure containing podocytes (examples highlighted by white arrows) connected to other tubular segments that are marked by aPKC but lack NPHS1. Nephrons are seen throughout the field and are packed together so that individual nephrons cannot be easily separated visually. MAFBmTagBFP2 reporter is expressed specifically in NPHS1 expressing podocytes, but is absent from other nephron segments (aPKC+, NPHS1− regions) or from other cell types. Images are maximum projections (50 µm span). e,f. Both conditions have a similar axial morphology in nephron-containing regions when viewed as an orthogonal slice (ie along the imaging Z-axis). A single orthogonal slice rendered from a 3D stack is shown. g,h. Cropped high-resolution fields showing a single glomerulus for each condition confirm co-expression of MAFBmtagBFP2 reporter and NPHS1 in podocytes. A single confocal slice is shown. All images are representative of at least n = 3 stained samples.
Extended Data Fig. 5 Quantification of large image data sets associated with organoids used for single cell RNA seq.
Line organoids are approximately 12 mm long. a. Representative images from 3 separate wells across replicates and conditions. b. Quantification of MAFB-mTagBFP2 reporter area by set and condition. Data is as in Fig. 5b, but here is separated by set. c. Quantification of GATA3-mCherry reporter area. Note that Y-axis scale differs between B and c, as GATA3 area represents a substantially smaller proportion of the organoid in most cases. d. GATA3 area as a proportion of total measured reporter area (MAFB + GATA3), highlighting a shift in R0 toward a more distalised fate. E. The total number of individual organoids used for quantification, by set and condition.
Extended Data Fig. 6 Analysis of single cell RNA datasets.
a. Variability within the datasets represented as a UMAP plot, coloured by transcriptional cluster, predicted cell cycle phase, main cell type and organoid conformation (clockwise from top left). b. Marker genes of main cells type, WT1 and PAX2 (nephron), PDGFRA (stroma) and SOX17 (endothelial). c. Proportion of each cell type in replicate conditions. P value (one-way ANOVA) indicated if p < 0.2. D. UMAP representation of nephron cells after re-transformation and clustering at higher resolution. Plots are coloured by transcriptional cluster, predicted cell cycle phase and organoid conformation. Cluster identities are stated. e. Marker genes identifying each cluster: GATA3 (distal), HNF1B (pre-tubule), CUBN (proximal), HNF4A (proximal), FOXC2 (pre-podocyte), MAFB (pre-podocyte / podocyte), PODXL (podocyte), SIX2 (progenitor), EYA1 (progenitor). f. Stromal UMAP coloured by transcriptional cluster, predicted cell cycle phase and organoid conformation (top to bottom). g. Markers of specific stromal clusters; SIX2, LYPD1, FOXC2, HOXA11 (Cluster 3, nephron progenitor-like), WNT5A, LHX9 (Cluster 7) and ZIC1 and ZIC4 (Cluster 10). h. Heatmap of scaled log counts per million of pseudo bulk counts from scRNAseq sets for the top 100 most significantly expressed genes identified in bulk RNAseq analysis (Fig. 4). Each column represents a single cluster from a single replicate (for example R40, Nephron, Set 1). Hierarchical clustering of the limited gene set indicates that bulk-RNAseq changes are largely driven by changes in the nephrons and endothelial cells.
Extended Data Fig. 7 The spatial distribution of stromal markers by wholemount immunoflouresence.
a–c. Immunofluorescence staining for markers of organoid stromal populations based on scRNA profiling. R0 organoids consist of a nephron containing area (Nephrons), a central role (Core) where nephrons are largely absent, and a thin edge (Thin edge) of monolayer cells that are typically not observed in brightfield imaging. R40 line organoids are primarily composed of a dense nephron-containing region and a thin monolayer edge, with no central core. Stromal population markers (A) MEIS1/2/3, (B) SIX1 and (C) SOX9 are present in the areas surrounding nephrons, and within the thin monolayer sheet at the edge of each organoid, but are largely absent from the central core of R0 organoids. Representative images from n = 3 organoids stained per condition are shown. Images are maximum projections spanning the full volume of the organoid. d. UMAP plots representing stromal cells in scRNA datasets, colour coded to show expression of MEIS1, MEIS2, SIX1 and SOX9. These combined markers include most of the cells in the dataset, suggesting that the absence of staining in the central core observed in (E) may indicate low overall cellularity in that region.
Extended Data Fig. 8 Direct comparison between kidney organoids and human fetal kidney confirms improved maturation of proximal tubules within R40 bioprinted lines.
a. UMAP plots comparing transcriptional identity based on unbiased clustering in Seurat (left) and prediction using the scPred method where cells are according to their similarity to a human fetal kidney (HFK) dataset (right). Identity assignment is based the most similar human fetal kidney cell type. b. The proportion of cells assigned to each cell type identity across replicates. Points show individual replicate values colour coded by replicate barcode (where HTO-1 is Set 1). Bars show SEM. P-values based on one-way ANOVA indicate a significant difference in the number of cells predicted to be Pre-Pod cells, with greatest abundance in the R40 datasets. Bioprinted conditions (R40 and R0) have more cells predicted to be podocytes, and less distal and pre-tubule cells. However, these changes were not significant. These results support the trends presented in Fig. 5. c. The distribution of maximum similarity scores for the classification of each cell across conformations, plotted by cell type predicted. Most cells show a high similarity to the predicted fetal kidney cell type. d. Genes identified as significantly increased in R40 versus Manual organoids (SLC51B, FABP3 and SULT1E1) are expressed in the mature proximal tubule cells of human fetal kidney, confirming their association with a more mature cell type. A gene that was significantly decreased in R40 vs Manual organoids (SPP1) is expressed selectively in less mature cell types, supporting increased maturity in R40 proximal cells. UMAP shows transcriptional identity in human fetal kidney data. Top left plot is colour coded by human fetal kidney cell types specific to developing (renal vesicle and comma shaped body [RV_CSB], blue; proximal early nephron [PEN], red) and mature proximal tubule (PT, green). Lower left plot shows a ‘dot plot’ style representation of selected gene where size indicates the percentage of HFK cells expressing the gene and colour indicates normalised expression level. Normalised expression of each gene per cell is indicated on individual UMAP plots where expression is colour coded.
Supplementary information
Supplementary Information
Tables 1 and 6, Methods and references.
Supplementary Data 1
Differentially expressed genes between conditions, within clusters for nephron and stromal subsets of scRNA data. Data for each comparison are contained within separate tabs, with a guide to set naming in the first tab.
Supplementary Table 2
Differentially expressed genes identified between ratio 40 and ratio 0 organoids, identified in bulk-RNAseq. Positive log fold-change indicates increased expression in R40
Supplementary Table 3
Whole dataset cluster markers for integrated scRNAseq dataset
Supplementary Table 4
Nephron cluster markers for integrated scRNAseq dataset
Supplemetnary Table 5
Stromal cluster markers for integrated scRNAseq dataset
Supplementary Video 1
Novogen 3D bioprinter generating kidney organoids.
Supplementary Video 2
3D rendering of a bioprinted dot organoid from Fig. 4d.
Supplementary Video 2
3D rendering of a bioprinted line organoid from Fig. 4..
Rights and permissions
About this article
Cite this article
Lawlor, K.T., Vanslambrouck, J.M., Higgins, J.W. et al. Cellular extrusion bioprinting improves kidney organoid reproducibility and conformation. Nat. Mater. 20, 260–271 (2021). https://doi.org/10.1038/s41563-020-00853-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-020-00853-9
This article is cited by
-
Role of biophysics and mechanobiology in podocyte physiology
Nature Reviews Nephrology (2024)
-
Harnessing 3D in vitro systems to model immune responses to solid tumours: a step towards improving and creating personalized immunotherapies
Nature Reviews Immunology (2024)
-
Advances in pediatric acute kidney injury pathobiology: a report from the 26th Acute Disease Quality Initiative (ADQI) conference
Pediatric Nephrology (2024)
-
From Pluripotent Stem Cells to Organoids and Bioprinting: Recent Advances in Dental Epithelium and Ameloblast Models to Study Tooth Biology and Regeneration
Stem Cell Reviews and Reports (2024)
-
HIF-1α promotes kidney organoid vascularization and applications in disease modeling
Stem Cell Research & Therapy (2023)