Abstract
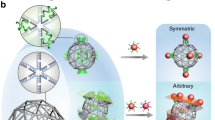
The ability to organize nanoscale objects into well-defined three-dimensional (3D) arrays can translate advances in nanoscale synthesis into targeted material fabrication. Despite successes in nanoparticle assembly, most extant methods are system specific and not fully compatible with biomolecules. Here, we report a platform for creating distinct 3D ordered arrays from different nanomaterials using DNA-prescribed and valence-controlled material voxels. These material voxels consist of 3D DNA frames that integrate nano-objects within their scaffold, thus enabling the object’s valence and coordination to be determined by the frame’s vertices, which can bind to each other through hybridization. Such DNA material voxels define the lattice symmetry through the spatially prescribed valence decoupling the 3D assembly process from the nature of the nanocomponents, such as their intrinsic properties and shapes. We show this by assembling metallic and semiconductor nanoparticles and also protein superlattices. We support the technological potential of such an assembly approach by fabricating light-emitting 3D arrays with diffraction-limited spectral purity and 3D enzymatic arrays with increased activity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within this article and its Supplementary Information, or from the corresponding author on reasonable request.
Code availability
The scripts used in X-ray scattering analysis and modelling are a part of the ScatterSim software package, a Python package available for download through GitHub (https://github.com/CFN-softbio/scattersim) or from the corresponding author on reasonable request.
References
Jones, M. R. et al. DNA-nanoparticle superlattices formed from anisotropic building blocks. Nat. Mater. 9, 913–917 (2010).
Lu, F., Yager, K. G., Zhang, Y. G., Xin, H. L. & Gang, O. Superlattices assembled through shape-induced directional binding. Nat. Commun. 6, 6912 (2015).
Agarwal, U. & Escobedo, F. A. Mesophase behaviour of polyhedral particles. Nat. Mater. 10, 230–235 (2011).
Gantapara, A. P., de Graaf, J., van Roij, R. & Dijkstra, M. Phase diagram and structural diversity of a family of truncated cubes: degenerate close-packed structures and vacancy-rich states. Phys. Rev. Lett. 111, 015501 (2013).
Talapin, D. V. et al. Quasicrystalline order in self-assembled binary nanoparticle superlattices. Nature 461, 964–967 (2009).
Damasceno, P. F., Engel, M. & Glotzer, S. C. Predictive self-assembly of polyhedra into complex structures. Science 337, 453–457 (2012).
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982).
Chen, J. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
He, Y. et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 452, 198–201 (2008).
Seeman, N. C. DNA in a material world. Nature 421, 427–431 (2003).
Halverson, J. D. & Tkachenko, A. V. DNA-programmed mesoscopic architecture. Phys. Rev. E 87, 062310 (2013).
Zheng, J. et al. Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 6, 1502–1504 (2006).
Nykypanchuk, D., Maye, M. M., van der Lelie, D. & Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 451, 549–552 (2008).
Park, S. Y. et al. DNA-programmable nanoparticle crystallization. Nature 451, 553–556 (2008).
Vo, T. et al. Stoichiometric control of DNA-grafted colloid self-assembly. Proc. Natl Acad. Sci. USA 112, 4982–4987 (2015).
Niemeyer, C. M., Koehler, J. & Wuerdemann, C. DNA-directed assembly of bienzymic complexes from in vivo biotinylated NAD(P)H:FMN oxidoreductase and luciferase. ChemBioChem 3, 242–245 (2002).
Lee, J. B. et al. A mechanical metamaterial made from a DNA hydrogel. Nat. Nanotechnol. 7, 816–820 (2012).
Church, G. M., Gao, Y. & Kosuri, S. Next-generation digital information storage in DNA. Science 337, 1628 (2012).
Zheng, J. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009).
Wang, X. et al. An organic semiconductor organized into 3D DNA arrays by ‘bottom-up’ rational design. Angew. Chem. Int. Ed. 56, 6445–6448 (2017).
Hao, Y. et al. A device that operates within within a self-assembled 3D DNA crystal. Nat. Chem. 9, 824–827 (2017).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Zhang, T. et al. 3D DNA origami crystals. Adv. Mater. 30, 1800273 (2018).
Yager, K. G., Zhang, Y. G., Lu, F. & Gang, O. Periodic lattices of arbitrary nano-objects: modeling and applications for self-assembled systems. J. Appl. Crystallogr. 47, 118–129 (2014).
Tian, Y. et al. Lattice engineering through nanoparticle–DNA frameworks. Nat. Mater. 15, 654–661 (2016).
Liu, W. et al. Diamond family of nanoparticle superlattices. Science 351, 582–586 (2016).
Macfarlane, R. J. et al. Nanoparticle superlattice engineering with DNA. Science 334, 204–208 (2011).
Rogers, W. B. & Crocker, J. C. Direct measurements of DNA-mediated colloidal interactions and their quantitative modeling. Proc. Natl Acad. Sci. USA 108, 15687–15692 (2011).
Varilly, P., Angioletti-Uberti, S., Mognetti, B. M. & Frenkel, D. A general theory of DNA-mediated and other valence-limited colloidal interactions. J. Chem. Phys. 137, 094108 (2012).
Travesset, A. Binary nanoparticle superlattices of softparticle systems. Proc. Natl Acad. Sci. USA 112, 9563–9567 (2015).
Wertheim, M. S. Fluids with highly directional attractive forces. iii. multiple attraction sites. J. Stat. Phys. 42, 459–476 (1986).
Damasecno, P. F., Engel, M. & Glotzer, S. C. Crystalline assemblies and densest packings of a family of truncated tetrahedra and the role of directional entropic forces. ACS Nano 6, 609–614 (2012).
Murray, C. B., Kagan, C. R. & Bawendi, M. G. Synthesis and characterization of monodisperse nanocrystals and close-packed nanocrystal assemblies. Annu. Rev. Mater. Sci. 30, 545–610 (2000).
Oliver, J. Quantum dots: global market growth and future commercial prospects. BCC Res. 5, 1–4 (2016).
Jang, E. et al. White-light-emitting diodes with quantum dot color converters for display backlights. Adv. Mater. 22, 3076–3080 (2010).
Kim, T.-H. et al. Full-colour quantum dot displays fabricated by transfer printing. Nat. Photonics 5, 176–182 (2011).
Wood, V. & Bulović, V. Colloidal quantum dot light-emitting devices. Nano Rev. 1, 5202 (2010).
Chang, T.-W. F. et al. High near-infrared photoluminescence quantum efficiency from PbS nanocrystals in polymer films. Synth. Met. 148, 257–261 (2005).
Kagan, C. R., Murray, C. B., Nirmal, M. & Bawendi, M. G. Electronic energy transfer in CdSe quantum dot solids. Phys. Rev. Lett. 76, 1517–1520 (1996).
Baimuratov, A. S., Rukhlenko, I. D., Turkov, V. K., Baranov, A. V. & Fedorov, A. V. Quantum-dot supercrystals for future nanophotonics. Sci. Rep. 3, 1727 (2013).
Kim, D. et al. Evidence of quantum resonance in periodically-ordered three-dimensional superlattice of CdTe quantum dots. Nano Lett. 15, 4343–4347 (2015).
Wilner, O. I. et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat. Nanotechnol. 4, 249–249 (2009).
Fu, J., Liu, M., Liu, Y., Woodbury, N. W. & Yan, H. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc. 134, 5516–5519 (2012).
Zhao, Z. et al. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 7, 10619 (2016).
Acknowledgements
We thank W. Xia for help with cryo-STEM imaging, M. Ji for help with negative stained TEM imaging, Y. Zhang and A. Fluerasu for help with SAXS measurement at the CHX beamline (NSLS-II at the Brookhaven National Laboratory), M. Cotlet and J. Chen for help with optical characterization, and D. Chen for help with illustrations. Cryo-EM data were collected at the David Van Andel Advanced Cryo-Electron Microscopy Suite in the Van Andel Research Institute. Y.T.’s work at Nanjing University was supported by Jiangsu Youth Fund of China (grant no. BK20180337) and the Fundamental Research Funds for the Central Universities (grant no. 14380151). H.L. was supported by the US National Institutes of Health (grant nos. GM111472 and GM124170) and the Van Andel Research Institute. This research used resources of the Center for Functional Nanomaterials, and the National Synchrotron Light Source II, which are US DOE Office of Science Facilities at Brookhaven National Laboratory under contract no. DE-SC0012704. This work was supported by the US Department of Energy, Office of Basic Energy Sciences, grant no. DE-SC0008772.
Author information
Authors and Affiliations
Contributions
Y.T. and O.G. conceived and designed the experiments. Y.T. performed the assembly experiments. J.R.L. and K.G.Y. contributed to the model fitting of the SAXS data. Y.T., L.B. and H.L. contributed to the cryo-EM imaging and reconstruction. Y.T. and H.L.X. contributed to the electron microscopy imaging of the superlattice. T.V. and S.K.K. performed computational modelling. J.S.K., B.M. and Y.X. prepared optical and enzymatic samples and conducted the experiments. R.L. and M.F. helped with SAXS experiments. Y.T. and O.G. wrote the paper. O.G. supervised the project. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Sections 1–8, Figs. 1–61, Tables 1–3 and references.
Rights and permissions
About this article
Cite this article
Tian, Y., Lhermitte, J.R., Bai, L. et al. Ordered three-dimensional nanomaterials using DNA-prescribed and valence-controlled material voxels. Nat. Mater. 19, 789–796 (2020). https://doi.org/10.1038/s41563-019-0550-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-019-0550-x
This article is cited by
-
Exploring the diverse biomedical applications of programmable and multifunctional DNA nanomaterials
Journal of Nanobiotechnology (2023)
-
Genetically encoded protein crystals by hierarchical design
Nature Materials (2023)
-
Fabricating higher-order functional DNA origami structures to reveal biological processes at multiple scales
NPG Asia Materials (2023)
-
Accurate computational design of three-dimensional protein crystals
Nature Materials (2023)
-
Interfacing DNA nanotechnology and biomimetic photonic complexes: advances and prospects in energy and biomedicine
Journal of Nanobiotechnology (2022)