Abstract
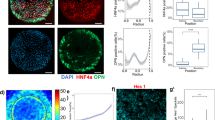
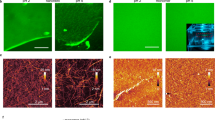
Cell size and shape affect cellular processes such as cell survival, growth and differentiation1,2,3,4, thus establishing cell geometry as a fundamental regulator of cell physiology. The contributions of the cytoskeleton, specifically actomyosin tension, to these effects have been described, but the exact biophysical mechanisms that translate changes in cell geometry to changes in cell behaviour remain mostly unresolved. Using a variety of innovative materials techniques, we demonstrate that the nanostructure and lipid assembly within the cell plasma membrane are regulated by cell geometry in a ligand-independent manner. These biophysical changes trigger signalling events involving the serine/threonine kinase Akt/protein kinase B (PKB) that direct cell-geometry-dependent mesenchymal stem cell differentiation. Our study defines a central regulatory role by plasma membrane ordered lipid raft microdomains in modulating stem cell differentiation with potential translational applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chen, C. S., Mrksich, M., Huang, S., Whitesides, G. M. & Ingber, D. E. Geometric control of cell life and death. Science 276, 1425–1428 (1997).
Kilian, Ka, Bugarija, B., Lahn, B. T. & Mrksich, M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl Acad. Sci. USA 107, 4872–4877 (2010).
McBeath, R., Pirone, D. M., Nelson, C. M., Bhadriraju, K. & Chen, C. S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6, 483–495 (2004).
Aragona, M. et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059 (2013).
Head, B. P., Patel, H. H. & Insel, P. A. Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim. Biophys. Acta 1838, 532–545 (2014).
Simons, K. & Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39 (2000).
Liu, A. P. & Fletcher, D. A. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys. J. 91, 4064–4070 (2006).
Gaus, K., Le Lay, S., Balasubramanian, N. & Schwartz, M. Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 174, 725–734 (2006).
Head, B. P. et al. Microtubules and actin microfilaments regulate lipid raft/caveolae localization of adenylyl cyclase signaling components. J. Biol. Chem. 281, 26391–26399 (2006).
Lingwood, D. & Simons, K. Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010).
Head, B. P. & Insel, P. A. Do caveolins regulate cells by actions outside of caveolae? Trends Cell Biol. 17, 51–57 (2007).
Palazzo, A. F., Eng, C. H., Schlaepfer, D. D., Marcantonio, E. E. & Gundersen, G. G. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science 303, 836–839 (2004).
Kamiguchi, H. The region-specific activities of lipid rafts during axon growth and guidance. J. Neurochem. 98, 330–335 (2006).
Blank, N. et al. Cholera toxin binds to lipid rafts but has a limited specificity for ganglioside GM1. Immunol. Cell Biol. 85, 378–382 (2007).
Wüstner, D. Fluorescent sterols as tools in membrane biophysics and cell biology. Chem. Phys. Lipids 146, 1–25 (2007).
Parton, R. G. & Simons, K. The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol. 8, 185–194 (2007).
Sezgin, E. et al. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 7, 1042–1051 (2012).
Gao, X. & Zhang, J. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol. Biol. Cell 19, 4366–4373 (2008).
Lasserre, R. et al. Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat. Chem. Biol. 4, 538–547 (2008).
Gao, X. et al. PI3K/Akt signaling requires spatial compartmentalization in plasma membrane microdomains. Proc. Natl Acad. Sci. USA 108, 14509–14514 (2011).
Calay, D. et al. Inhibition of Akt signaling by exclusion from lipid rafts in normal and transformed epidermal keratinocytes. J. Investigative Dermatol. 130, 1136–1145 (2010).
Manning, B. D. & Cantley, L. C. AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 (2007).
Schnitzer, J. E., Oh, P., Pinney, E. & Allard, J. Filipin-sensitive caveolae-mediated transport in endothelium: reduced transcytosis, scavenger endocytosis, and capillary permeability of select macromolecules. J. Cell Biol. 127, 1217–1232 (1994).
Hirai, H. et al. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol. Cancer Therapeutics 9, 1956–1967 (2010).
Vanhaesebroeck, B., Stephens, L. & Hawkins, P. PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell Biol. 13, 195–203 (2012).
Müller, P., Langenbach, A., Kaminski, A. & Rychly, J. Modulating the actin cytoskeleton affects mechanically induced signal transduction and differentiation in mesenchymal stem cells. PLoS ONE 8, 1–8 (2013).
Stevens, M. M. & George, J. H. Exploring and engineering the cell surface interface. Science 310, 1135–1138 (2005).
Place, E. S., Evans, N. D. & Stevens, M. M. Complexity in biomaterials for tissue engineering. Nat. Mater. 8, 457–470 (2009).
von Erlach, T. C., Hedegaard, M. A. B. & Stevens, M. M. High resolution Raman spectroscopy mapping of stem cell micropatterns. Analyst 140, 1798–1803 (2015).
Bertazzo, S., von Erlach, T., Goldoni, S., Çandarlıoğlu, P. L. & Stevens, M. M. Correlative light-ion microscopy for biological applications. Nanoscale 4, 2851–2854 (2012).
Tan, J. L., Liu, W., Nelson, C. M., Raghavan, S. & Chen, C. S. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 10, 865–72 (2004).
Horejs, C. M. et al. Preventing tissue fibrosis by local biomaterials interfacing of specific cryptic extracellular matrix information. Nat. Commun. 8, 1–15 (2017).
Leight, J. L., Wozniak, M. A., Chen, S., Lynch, M. L. & Chen, C. S. Matrix rigidity regulates a switch between TGF-1-induced apoptosis and epithelial-mesenchymal transition. Mol. Biol. Cell 23, 781–791 (2012).
Harris, A. R. & Charras, G. T. Experimental validation of atomic force microscopy-based cell elasticity measurements. Nanotechnology 22, 1–10 (2011).
Herrmann, I. K. et al. Differentiating sepsis from non-infectious systemic inflammation based on microvesicle-bacteria aggregation. Nanoscale 7, 13511–13520 (2015).
Acknowledgements
We thank H. M. Textor and F. Anderegg (ETH Zurich) for providing silicon masters for micro-contact printing as well as S. Rothery for training and guidance regarding TIRF microscopy (FILM facility at Imperial College London). T.C.v.E. was supported by an EPSRC DTA PhD award. S.B. was supported by the Rosetrees Trust and the Stoneygate Trust and the Junior Research Fellowship scheme at Imperial College London. M.M.S. gratefully acknowledges ERC starting grant “Naturale” for funding (206807), Wellcome Trust Senior Investigator Award (098411/Z/12/Z) and the Rosetrees Trust. A.D.R.H. gratefully acknowledges ERC starting grant "ForceRegulation' (282051).
Author information
Authors and Affiliations
Contributions
T.C.v.E. designed experiments, developed the substrates and conducted experiments, analysed and interpreted the data and wrote the manuscript. S.B. designed and carried out ion and electron microscopy experiments and analysed the data. M.A.W. conducted viral transfection experiments and revised the manuscript. C.K. performed 3D plasma membrane reconstruction and analysis. B.K.R. and S.A. conducted AFM measurements. C.-M.H. carried out western blots. C.S.C. revised the manuscript and consulted in experimental design. A.D.R.H. revised the manuscript and supervised S.A. H.A. helped with hMSC cultivation and differentiation experiments and revised the manuscript. S.A.M. helped with cell micropattern preparations and revised the manuscript. S.G. supervised the project, helped in experimental design, data analysis and interpretation, and co-wrote the manuscript. M.M.S. supervised the project, co-wrote the manuscript and helped in experimental design and data interpretation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures.
Rights and permissions
About this article
Cite this article
von Erlach, T.C., Bertazzo, S., Wozniak, M.A. et al. Cell-geometry-dependent changes in plasma membrane order direct stem cell signalling and fate. Nature Mater 17, 237–242 (2018). https://doi.org/10.1038/s41563-017-0014-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-017-0014-0
This article is cited by
-
Nature-inspired micropatterns
Nature Reviews Methods Primers (2023)
-
Regulation of biological processes by intrinsically chiral engineered materials
Nature Reviews Materials (2023)
-
Computer vision meets microfluidics: a label-free method for high-throughput cell analysis
Microsystems & Nanoengineering (2023)
-
How the mechanical microenvironment of stem cell growth affects their differentiation: a review
Stem Cell Research & Therapy (2022)
-
Role of actin cytoskeleton in cargo delivery mediated by vertically aligned silicon nanotubes
Journal of Nanobiotechnology (2022)