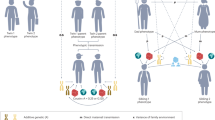
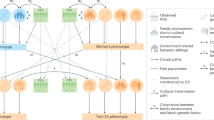
Abstract
Genome-wide association studies on human behavioural traits are producing large amounts of polygenic signals with significant predictive power and potentially useful biological clues. Behavioural traits are more distal and are less directly under biological control compared with physical characteristics, which makes the associated genetic effects harder to interpret. The results of genome-wide association studies for human behaviour are likely made up of a composite of signals from different sources. While sample sizes continue to increase, we outline additional steps that need to be taken to better delineate the origin of the increasingly stronger polygenic signals. In addition to genetic effects on the traits themselves, the major sources of polygenic signals are those that are associated with correlated traits, environmental effects and ascertainment bias. Advances in statistical approaches that disentangle polygenic effects from different traits as well as extending data collection to families and social circles with better geographical coverage will probably contribute to filling the gap of knowledge between genetic effects and behavioural outcomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Boomsma, D., Busjahn, A. & Peltonen, L. Classical twin studies and beyond. Nat. Rev. Genet. 3, 872–882 (2002).
Polderman, T. J. et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 47, 702–709 (2015).
Turkheimer, E. Three laws of behavior genetics and what they mean. Curr. Dir. Psychol. Sci. 9, 160–164 (2000).
Gusella, J. F. et al. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature 306, 234–238 (1983).
Tsui, L.-C. et al. Cystic fibrosis locus defined by a genetically linked polymorphic DNA marker. Science 230, 1054–1057 (1985).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Daly, M. J., Rioux, J. D., Schaffner, S. F., Hudson, T. J. & Lander, E. S. High-resolution haplotype structure in the human genome. Nat. Genet. 29, 229–232 (2001).
Gabriel, S. B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
The International HapMap Consortium The international HapMap project. Nature 426, 789–796 (2003).
DeWan, A. et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 314, 989–992 (2006).
The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Klein, R. J. et al. Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389 (2005).
Li, Y., Willer, C., Sanna, S. & Abecasis, G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 10, 387–406 (2009).
Duncan, L. E., Ostacher, M. & Ballon, J. How genome-wide association studies (GWAS) made traditional candidate gene studies obsolete. Neuropsychopharmacology 44, 1518–1523 (2019).
Border, R. et al. No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am. J. Psychiatry 176, 376–387 (2019).
Morris, A. P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012).
Locke, A. E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015).
Ehret, G. B. et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478, 103–109 (2011).
Lango, H. A. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Yang, J. et al. FTO genotype is associated with phenotypic variability of body mass index. Nature 490, 267–272 (2012).
Ripke, S. et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Ripke, S. et al. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 43, 969–976 (2011).
Stahl, E. A. et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019).
Sklar, P. et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43, 977–983 (2011).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Ripke, S. et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18, 497–511 (2013).
Lo, M.-T. et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat. Genet. 49, 152–156 (2017).
Ganna, A. et al. Large-scale GWAS reveals insights into the genetic architecture of same-sex sexual behavior. Science 365, eaat7693 (2019).
Liu, M., Jiang, Y. & Wedow, R. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244 (2019).
Pasman, J. A. et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal effect of schizophrenia liability. Nat. Neurosci. 21, 1161–1170 (2018).
Sniekers, S. et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat. Genet. 49, 1107–1112 (2017).
Lee, J. J. et al. Gene discovery and polygenic prediction from a 1.1-million-person GWAS of educational attainment. Nat. Genet. 50, 1112–1121 (2018).
Visscher, P. M., Brown, M. A., McCarthy, M. I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Visscher, P. M. et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017).
Fisher, R. A. XV.—The correlation between relatives on the supposition of Mendelian inheritance. Earth Environ. Sci. Trans. R. Soc. Edinb. 52, 399–433 (1919).
Hivert, V. et al. Estimation of non-additive genetic variance in human complex traits from a large sample of unrelated individuals. Am. J. Hum. Genet. https://doi.org/10.1016/j.ajhg.2021.02.014 (2021).
Hill, W. G., Goddard, M. E. & Visscher, P. M. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4, e1000008 (2008).
Crow, J. F. On epistasis: why it is unimportant in polygenic directional selection. Philos. Trans. R. Soc. B 365, 1241–1244 (2010).
Wainschtein, P. et al. Recovery of trait heritability from whole genome sequence data. Preprint at bioRxiv https://doi.org/10.1101/588020 (2019).
Kaiser, J. ‘Landmark’ study resolves a major mystery of how genes govern human height. Science (3 November 2020).
Nelson, M. R. et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860 (2015).
King, E. A., Davis, J. W. & Degner, J. F. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 15, e1008489 (2019).
Sekar, A. et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 (2016).
Ward, L. D. & Kellis, M. Interpreting noncoding genetic variation in complex traits and human disease. Nat. Biotechnol. 30, 1095–1106 (2012).
Emes, R. D. et al. Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat. Neurosci. 11, 799–806 (2008).
Ip, H. F. et al. Characterizing the relation between expression QTLs and complex traits: exploring the role of tissue specificity. Behav. Genet. 48, 374–385 (2018).
Qi, T. et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 9, 2282 (2018).
Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Koopmans, F. et al. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103, 217–234 (2019).
Torkamani, A., Wineinger, N. E. & Topol, E. J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590 (2018).
Ikeda, M., Saito, T., Kanazawa, T. & Iwata, N. Polygenic risk score as clinical utility in psychiatry: a clinical viewpoint. J. Hum. Genet. 66, 53–60 (2020).
Wray, N. R. et al. From basic science to clinical application of polygenic risk scores: a primer. JAMA Psychiatry 78, 101–109 (2021).
The International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
The Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke, S., Walters, J. T. R. & O’Donovan, M. C. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. Preprint at medRxiv https://doi.org/10.1101/2020.09.12.20192922 (2020).
Rietveld, C. A. et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340, 1467–1471 (2013).
Wray, N. R. et al. Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet. 14, 507–515 (2013).
Lambert, S. A., Abraham, G. & Inouye, M. Towards clinical utility of polygenic risk scores. Hum. Mol. Genet. 28, R133–R142 (2019).
Maas, P. et al. Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States. JAMA Oncol. 2, 1295–1302 (2016).
Schumacher, F. R. et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 50, 928–936 (2018).
Sharp, S. A. & Rich, S. S. Development and standardization of an improved type 1 diabetes genetic risk score for use in newborn screening and incident diagnosis. Diabetes Care 42, 200–207 (2019).
Sparano, J. A. et al. Clinical and genomic risk to guide the use of adjuvant therapy for breast cancer. N. Engl. J. Med. 380, 2395–2405 (2019).
Inouye, M. et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J. Am. Coll. Cardiol. 72, 1883–1893 (2018).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Abdellaoui, A. et al. Population structure, migration, and diversifying selection in the Netherlands. Eur. J. Hum. Genet. 21, 1277–1285 (2013).
Kerminen, S. et al. Fine-scale genetic structure in Finland. G3 7, 3459–3468 (2017).
Novembre, J. et al. Genes mirror geography within Europe. Nature 456, 98–101 (2008).
Leslie, S. et al. The fine-scale genetic structure of the British population. Nature 519, 309–314 (2015).
Price, A. L., Zaitlen, N. A., Reich, D. & Patterson, N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11, 459–463 (2010).
Berg, J. J. et al. Reduced signal for polygenic adaptation of height in UK Biobank. eLife 8, e39725 (2019).
Cardon, L. R. & Palmer, L. J. Population stratification and spurious allelic association. Lancet 361, 598–604 (2003).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Galton, F. Typical laws of heredity. III. Nature 15, 512–514 (1877).
Galton, F. I. Co-relations and their measurement, chiefly from anthropometric data. Proc. R. Soc. Lond. 45, 135–145 (1889).
Plana-Ripoll, O. et al. Exploring comorbidity within mental disorders among a Danish national population. JAMA Psychiatry 76, 259–270 (2019).
Momen, N. C. et al. Association between mental disorders and subsequent medical conditions. N. Engl. J. Med. 382, 1721–1731 (2020).
Polderman, T. J. et al. A genetic study on attention problems and academic skills: results of a longitudinal study in twins. J. Canadian Acad. Child Adolesc. Psychiatry 20, 22–34 (2011).
Cardno, A. G., Rijsdijk, F. V., Sham, P. C., Murray, R. M. & McGuffin, P. A twin study of genetic relationships between psychotic symptoms. Am. J. Psychiatry 159, 539–545 (2002).
Polderman, T., Hoekstra, R., Posthuma, D. & Larsson, H. The co-occurrence of autistic and ADHD dimensions in adults: an etiological study in 17 770 twins. Transl. Psychiatry 4, e435–e435 (2014).
Bartels, M. et al. The five factor model of personality and intelligence: a twin study on the relationship between the two constructs. Pers. Individ. Dif. 53, 368–373 (2012).
Plomin, R. & DeFries, J. Multivariate behavioral genetic analysis of twin data on scholastic abilities. Behav. Genet. 9, 505–517 (1979).
Verweij, K. J., Huizink, A. C., Agrawal, A., Martin, N. G. & Lynskey, M. T. Is the relationship between early-onset cannabis use and educational attainment causal or due to common liability? Drug Alcohol Depend. 133, 580–586 (2013).
Zietsch, B., Verweij, K., Bailey, J., Wright, M. & Martin, N. Genetic and environmental influences on risky sexual behaviour and its relationship with personality. Behav. Genet. 40, 12–21 (2010).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Watanabe, K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 51, 1339–1348 (2019).
van Rheenen, W., Peyrot, W. J., Schork, A. J., Lee, S. H. & Wray, N. R. Genetic correlations of polygenic disease traits: from theory to practice. Nat. Rev. Genet. 20, 567–581 (2019).
Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, 224 (2018).
Verbanck, M., Chen, C.-y, Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Keller, M. C. et al. The genetic correlation between height and IQ: shared genes or assortative mating? PLoS Genet. 9, e1003451 (2013).
Hugh-Jones, D., Verweij, K. J. H., Pourcain, B. S. & Abdellaoui, A. Assortative mating on educational attainment leads to genetic spousal resemblance for polygenic scores. Intelligence 59, 103–108 (2016).
Robinson, M. R. et al. Genetic evidence of assortative mating in humans. Nat. Hum. Behav. 1, 0016 (2017).
Kemper, K. E. et al. Phenotypic covariance across the entire spectrum of relatedness for 86 billion pairs of individuals. Nat. Commun. 12, 1050 (2021).
Turley, P. et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 50, 229–237 (2018).
Baselmans, B. M. et al. Multivariate genome-wide analyses of the well-being spectrum. Nat. Genet. 51, 445–451 (2019).
Lee, P. H. et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482 (2019).
Hill, W. et al. A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol. Psychiatry 24, 169–181 (2019).
Peyrot, W. J. & Price, A. L. Identifying loci with different allele frequencies among cases of eight psychiatric disorders using CC-GWAS. Nat. Genet. 53, 445–454 (2021).
Anttila, V. et al. Analysis of shared heritability in common disorders of the brain. Science 360, eaap8757 (2018).
Lee, P. H. et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482 (2019).
Grotzinger, A. D. et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav. 3, 513–525 (2019).
Demange, P. A. et al. Investigating the genetic architecture of noncognitive skills using GWAS-by-subtraction. Nat. Genet. 53, 35–44 (2021).
Marees, A. T. et al. Genetic correlates of socio-economic status influence the pattern of shared heritability across mental health traits. Nat. Hum. Behav. https://doi.org/10.1038/s41562-021-01053-4 (2021).
Lawlor, D. A., Harbord, R. M., Sterne, J. A., Timpson, N. & Davey Smith, G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27, 1133–1163 (2008).
Munafò, M. R. et al. Association between genetic variants on chromosome 15q25 locus and objective measures of tobacco exposure. J. Natl Cancer Inst. 104, 740–748 (2012).
Voight, B. F. et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet 380, 572–580 (2012).
Rossouw, J. E. et al. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results from the Women’s Health Initiative randomized controlled trial. JAMA 288, 321–333 (2002).
Davey Smith, G. & Ebrahim, S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32, 1–22 (2003).
Abdellaoui, A. et al. Genetic correlates of social stratification in Great Britain. Nat. Hum. Behav. 3, 1332–1342 (2019).
Kong, A. et al. The nature of nurture: effects of parental genotypes. Science 359, 424–428 (2018).
Demange, P. A. et al. Parental influences on offspring education: indirect genetic effects of non-cognitive skills. Preprint at bioRxiv https://doi.org/10.1101/2020.09.15.296236 (2020).
Selzam, S. et al. Comparing within- and between-family polygenic score prediction. Am. J. Hum. Genet. 105, 351–363 (2019).
Cheesman, R. et al. Comparison of adopted and nonadopted individuals reveals gene–environment interplay for education in the UK Biobank. Psychol. Sci. 31, 582–591 (2020).
Howe, L. J. et al. Within-sibship GWAS improve estimates of direct genetic effects. Preprint at bioRxiv https://doi.org/10.1101/2021.03.05.433935 (2021).
Hur, Y.-M. & Craig, J. M. Twin registries worldwide: an important resource for scientific research. Twin Res. Hum. Genet. 16, 1–12 (2013).
Hur, Y.-M. et al. Twin family registries worldwide: an important resource for scientific research. Twin Res. Hum. Genet. 22, 427–437 (2019).
Abdellaoui, A., Verweij, K. J. H. & Nivard, M. G. Geographic confounding in genome-wide association studies. Preprint at bioRxiv https://doi.org/10.1101/2021.03.18.435971 (2021).
Martin, J. et al. Association of genetic risk for schizophrenia with nonparticipation over time in a population-based cohort study. Am. J. Epidemiol. 183, 1149–1158 (2016).
Munafò, M. R., Tilling, K., Taylor, A. E., Evans, D. M. & Davey Smith, G. Collider scope: when selection bias can substantially influence observed associations. Int. J. Epidemiol. 47, 226–235 (2018).
Batty, G. D., Gale, C. R., Kivimäki, M., Deary, I. J. & Bell, S. Comparison of risk factor associations in UK Biobank against representative, general population based studies with conventional response rates: prospective cohort study and individual participant meta-analysis. BMJ 368, m131 (2020).
Fry, A. et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am. J. Epidemiol. 186, 1026–1034 (2017).
Adams, M. J. et al. Factors associated with sharing e-mail information and mental health survey participation in large population cohorts. Int. J. Epidemiol. 49, 410–421 (2020).
Tyrrell, J. et al. Genetic predictors of participation in optional components of UK Biobank. Nat. Commun. 12, 886 (2021).
Pirastu, N. et al. Genetic analyses identify widespread sex-differential participation bias. Nat. Genet. https://doi.org/10.1038/s41588-021-00846-7 (2021).
Xue, A. et al. Genome-wide analyses of behavioural traits biased by misreports and longitudinal changes. Nat. Commun. 12, 20211 (2021).
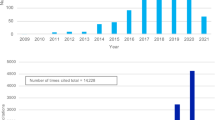
Mills, M. C. & Rahal, C. A scientometric review of genome-wide association studies. Commun. Biol. 2, 9 (2019).
Pedersen, C. B. et al. The iPSYCH2012 case–cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol. Psychiatry 23, 6–14 (2018).
Stefansson, K. Letters from Iceland. Nat. Genet. 47, 425 (2015).
Kerminen, S. et al. Geographic variation and bias in the polygenic scores of complex diseases and traits in Finland. Am. J. Hum. Genet. 104, 1169–1181 (2019).
Bulik-Sullivan, B. K., Loh, P. R., Finucane, H. K., Ripke, S. & Yang, J. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019).
Acknowledgements
A.A. and K.J.H.V. are supported by the Foundation Volksbond Rotterdam. A.A. is also supported by ZonMw grant no. 849200011 from The Netherlands Organisation for Health Research and Development.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Human Behaviour thanks Anders Børglum and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1: the numerical values of heritability estimates from Fig. 2, as well as the references for the twin/family studies and GWASs that Fig. 2 is based on.
Rights and permissions
About this article
Cite this article
Abdellaoui, A., Verweij, K.J.H. Dissecting polygenic signals from genome-wide association studies on human behaviour. Nat Hum Behav 5, 686–694 (2021). https://doi.org/10.1038/s41562-021-01110-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41562-021-01110-y
This article is cited by
-
Genetic influences and causal pathways shared between cannabis use disorder and other substance use traits
Molecular Psychiatry (2024)
-
Genetic predisposition for negative affect predicts mental health burden during the COVID-19 pandemic
European Archives of Psychiatry and Clinical Neuroscience (2024)
-
Multi-PGS enhances polygenic prediction by combining 937 polygenic scores
Nature Communications (2023)
-
Participation bias in the UK Biobank distorts genetic associations and downstream analyses
Nature Human Behaviour (2023)
-
Overlapping brain correlates of superior cognition among children at genetic risk for Alzheimer’s disease and/or major depressive disorder
Scientific Reports (2023)