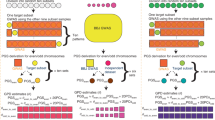
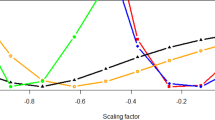
Abstract
Indirect genetic effects, the effects of the genotype of one individual on the phenotype of other individuals, are environmental factors associated with human disease and complex trait variation that could help to expand our understanding of the environment linked to complex traits. Here, we study indirect genetic effects in 80,889 human couples of European ancestry for 105 complex traits. Using a linear mixed model approach, we estimate partner indirect heritability and find evidence of partner heritability on ~50% of the analysed traits. Follow-up analysis suggests that in at least ~25% of these traits, the partner heritability is consistent with the existence of indirect genetic effects including a wide variety of traits such as dietary traits, mental health and disease. This shows that the environment linked to complex traits is partially explained by the genotype of other individuals and motivates the need to find new ways of studying the environment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data that support the results presented in this paper can be accessed from UK Biobank after publication. UK Biobank will link the dataset returned to the publication through their application system.
Code availability
The main results of this work were obtained using DISSECT19, which can be freely downloaded from http://www.dissect.ed.ac.uk.
References
Bijma, P. The quantitative genetics of indirect genetic effects: a selective review of modelling issues. Heredity 112, 61–69 (2014).
Wolf, J. B., Brodie, E. D. III, Cheverud, J. M., Moore, A. J. & Wade, M. J. Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. 13, 64–69 (1998).
Moore, A. J., Brodie, E. D. III & Wolf, J. B. Interacting phenotypes and the evolutionary process: I. Direct and indirect genetic effects of social interactions. Evolution 51, 1352–1362 (1997).
Mousseau, T. A. & Fox, C. W. The adaptive significance of maternal effects. Trends Ecol. Evol. 13, 403–407 (1998).
Willham, R. L. The covariance between relatives for characters composed of components contributed by related individuals. Biometrics 19, 18–27 (1963).
Santostefano, F., Wilson, A. J., Niemelä, P. T. & Dingemanse, N. J. Indirect genetic effects: a key component of the genetic architecture of behaviour. Sci. Rep. 7, 10235 (2017).
Brotherstone, S. et al. Competition effects in a young Sitka spruce (Picea sitchensis, Bong. Carr) clonal trial. Silvae Genet. 60, 149–155 (2011).
Camerlink, I., Ursinus, W. W., Bijma, P., Kemp, B. & Bolhuis, J. E. Indirect genetic effects for growth rate in domestic pigs alter aggressive and manipulative biting behaviour. Behav. Genet. 45, 117–126 (2015).
Warrington, N. M. et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat. Genet. 51, 804–814 (2019).
Kong, A. et al. The nature of nurture: effects of parental genotypes. Science 359, 424–428 (2018).
Domingue, B. W. et al. The social genome of friends and schoolmates in the national longitudinal study of adolescent to adult health. Proc. Natl Acad. Sci. USA 115, 702–707 (2018).
Baud, A., Casale, F. P., Nicod, J. & Stegle, O. Comparative architectures of direct and social genetic effects from the genome-wide association study of 170 phenotypes in laboratory mice. Preprint at bioRxiv https://doi.org/10.1101/302349 (2019).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Lee, S. H., Wray, N. R., Goddard, M. E. & Visscher, P. M. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 88, 294–305 (2011).
Dempster, E. R. & Lerner, I. M. Heritability of threshold characters. Genetics 35, 212–236 (1950).
Tenesa, A., Rawlik, K., Navarro, P. & Canela-Xandri, O. Genetic determination of height-mediated mate choice. Genome Biol. 16, 269 (2016).
Hugh-Jones, D., Verweij, K. J. H., St Pourcain, B. & Abdellaoui, A. Assortative mating on educational attainment leads to genetic spousal resemblance for polygenic scores. Intelligence 59, 103–108 (2016).
Stulp, G., Simons, M. J. P., Grasman, S. & Pollet, T. V. Assortative mating for human height: a meta-analysis. Am. J. Hum. Biol. 29, e22917 (2017).
Canela-Xandri, O., Law, A., Gray, A., Woolliams, J. A. & Tenesa, A. A new tool called DISSECT for analysing large genomic data sets using a big data approach. Nat. Commun. 6, 10162 (2015).
Fisher, R. A. Statistical Methods for Research Workers 4th edn (Oliver and Boyd, 1932).
Cheesman, R. et al. Comparison of adopted and non-adopted individuals reveals gene-environment interplay for education in the UK Biobank. Psychol. Sci. 31, 582–591 (2019).
Canela-Xandri, O., Rawlik, K. & Tenesa, A. An atlas of genetic associations in UK Biobank. Nat. Genet. 50, 1593–1599 (2018).
Bycroft, C. et al. Genome-wide genetic data on ~500,000 UK Biobank participants. Preprint at bioRxiv https://doi.org/10.1101/166298 (2017).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Visscher, P. M. A note on the asymptotic distribution of likelihood ratio tests to test variance components. Twin Res. Hum. Genet. 9, 490–495 (2006).
Sing, T., Sander, O., Beerenwinkel, N. & Lengauer, T. ROCR: visualizing classifier performance in R. Bioinformatics 21, 3940–3941 (2005).
Hanley, J. A. & McNeil, B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 (1982).
Acknowledgements
This research has been conducted using the UK Biobank Resource under project 788. The work was funded by Roslin Institute Strategic Programme Grants from the BBSRC (BBS/E/D/10002070 and BBS/E/D/30002275) and an MRC grant (MR/P015514/1). A.T. also acknowledges funding from the Medical Research Council Human Genetic Unit and Health Data Research UK (references HDR-9004 and HDR-9003). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Analyses were performed using the ARCHER UK National Supercomputing Service.
Author information
Authors and Affiliations
Contributions
C.X. contributed to the analysis. A.T. designed the study. C.X., O.C.-X., K.R. and A.T. contributed to the interpretation of data and the writing of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Human Behaviour thanks Loic Yengo, Piter Bijmaand and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Primary Handling Editor: Stavroula Kousta.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10, Supplementary Tables 12–21, Supplementary Methods, Supplementary Results and Supplementary References.
Supplementary Tables
Supplementary Tables 1–11.
Rights and permissions
About this article
Cite this article
Xia, C., Canela-Xandri, O., Rawlik, K. et al. Evidence of horizontal indirect genetic effects in humans. Nat Hum Behav 5, 399–406 (2021). https://doi.org/10.1038/s41562-020-00991-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41562-020-00991-9
This article is cited by
-
Indirect genetic effects are shaped by demographic history and ecology in Arabidopsis thaliana
Nature Ecology & Evolution (2023)
-
Partner choice, confounding and trait convergence all contribute to phenotypic partner similarity
Nature Human Behaviour (2023)
-
Partners in Health: Investigating Social Genetic Effects Among Married and Cohabiting Couples
Behavior Genetics (2023)
-
Correlations in sleeping patterns and circadian preference between spouses
Communications Biology (2023)
-
Dissecting indirect genetic effects from peers in laboratory mice
Genome Biology (2021)