Abstract
Variable trace metal concentrations in the Precambrian ocean were closely linked to oxygen availability, although less is known about the drivers of seawater trace metal chemistry after the spread of complex life into the Phanerozoic eon. A major phytoplankton succession took place at the transition from the Palaeozoic to the Mesozoic era (~250 Myr ago), from an ocean dominated by the green Archaeplastida to secondary endosymbiotic algae with red-algal-derived plastids. Here, our comparative genomic analysis of 26 complete proteomes and metal domain analysis of additional 608 partially complete sequences of phytoplankton reveal that groups with different evolutionary history have distinct metal-binding proteins and contrasting metal acquisition strategies, adapted to differing availability of trace metals. The secondary-endosymbiont-bearing lineages are better adapted to well-oxygenated, nutrient-poor environments. This is supported by an enhanced thiol-based binding affinity of their transporters, coupled with minimized proteomic requirement for trace elements such as iron, copper and zinc at both protein and domain levels. Such different metal requirements across these lineages suggest a drastic decline in open-ocean trace metal concentrations at the inception of the Mesozoic, contributing to the shifts in phytoplankton communities that drove major changes in ocean chemical buffering.
Similar content being viewed by others
Main
Trace element composition of the Precambrian ocean is of prime interest, due to the oxygen sensitivity of their availability, and is frequently inferred from the chemistry of indirect seawater recorders such as iron formations, black shale, pyrite records and carbonates1. Such an approach has rarely been applied to the Phanerozoic eon2,3, despite recent evidence of a transformation in the oxygen content of the upper ocean at the Palaeozoic/Mesozoic boundary4. Coincident with this change in oxygen, a Mesozoic algal revolution took place. Steranes in fossil records reveal that the Palaeozoic5 dominance of green algal phototrophs (Chlorophyta) ceased in the Mesozoic era (252–66 Myr ago; Ma) when the open ocean was taken over by the chlorophyll a+c-containing eukaryotes of the haptophyte and heterokont lineages whose plastids originate from red algae (Rhodophyta) via secondary endosymbiosis6,7,8 (Supplementary Fig. 1). These changing algal community structures and the development of mineralization strategies subsequently led to major changes in ocean ecology and chemical buffering9.
Recent molecular clock analyses indicate that the events underlying the evolutionary origin of algae, including the primary endosymbiosis and the subsequent spread of red plastids across the eukaryote tree, all pre-dated the ecological expansion of these groups by at least ~0.5–1 Gyr (ref. 5). This suggests that the selective pressures that underpinned the establishment of phytoplankton groups are distinct from those that drove their expansion5. Algal superfamilies are suspected to harbour distinct intracellular trace element stoichiometry10,11,12 highlighting potential differences in their physiological metal requirements for growth. To fulfil their needs for trace metals, phytoplankton have developed a complex network for metal uptake, chelation, transportation and storage, in which transmembrane metal transporters are crucial components13,14. Metal transport systems are key to dealing with metal limitation and toxicity in the environment, which underpins the environmental selection for different phytoplankton groups in the ocean. Episodes of transformation of phytoplankton domination in the ocean from the geological record, therefore, may provide evidence for change in seawater chemistry.
Here we employed a comparative genomic approach to analyse metal transporters and metal-binding proteins across a variety of phytoplankton species, abundant in the modern ocean, to understand whether patterns of trace metal usage and uptake are characteristic of different algal lineages (Figs. 1 and 2). The geological history of species succession in the ocean could then be used as a broad indicator of changing ocean trace metal chemistry, even though there may be some examples of decoupling between evolving marine chemistry and metal usage by phototrophs15.
a, Abundance of metalloproteins encoded in each species of phytoplankton (normalized to the proteome size). List 1–26: Synechococcus sp., Prochlorococcus marinus, Bathycoccus prasinos, Micromonas commode, Micromonas pusilla, Osetreococcus lucimarinus, Osetreococcus tauri, Auxenochlorella protothecoide, Coccomyxa subllipsoidea, Chlorella variabilis, Volvox carteri, Chlamydomonas reinhardtii, Monoraphidium neglectum, Klebsormidium flaccidum, Klebsormidium nitens, Chondrus crispus, Porphyra umbilicalis, Cyanidioschyzon merolae, Galdieria sulphuraria, Guillardia theta, Chrysochromulina sp., Emiliania huxleyi, Aurantiochytrium anophagefferens, Fragilariopsis cylindrus, Thalassiosira oceanica, Thalassiosira pseudonana. The dash lines separate different phytoplankton groups (cyanobacteria, primary endosymbionts, and secondary endosymbionts). b, Average abundance of metalloproteins in each group (normalized to proteome size: ncyanobacteria = 2, nprimary = 17, nsecondary = 7). c, Abundance of metal domains in different groups of phytoplankton (normalized to total number of protein domains: ncyanobacteria = 2, nprimary = 15, nsecondary = 11). d, Abundance of metal domains in primary endosymbionts (n = 64) versus secondary/tertiary endosymbionts (n = 544) from the MMETSP (normalized to total number of protein domains). Boxes mark the 25th and 75th percentiles of values for each group. The line in the middle of the box is the median. The whiskers show the maximum and minimum values of the group. The points show the outliers that exceed 1.5 × the interquartile range of the group. The numbers in b, c and d indicate the P values from the two-tailed t-test and are highlighted in bold when P < 0.05. For Fe, Zn, Cu and Ni, data from all three different analyses agree with each other. For Mo, the abundances of Mo-containing proteins (domains) are higher in eukaryotes than in prokaryotes. Data for Mn, Co and Cd do not fully agree in different analysis. There was no differentiation of the metal-binding domains from transporters and metalloproteins in metal domain analysis for c and d, and therefore the data in c and d are a mix of both, which may obscure the inference of metal use alone (Supplementary Discussion). Major functions of essential trace elements are shown in Supplementary Fig. 2.
a, The total number of genes in each species. Black bars represent the numbers of genes that have been assigned in the orthogroups, and the grey bars represent the unique genes found in each species. b, The numbers of different transporters found in each species. c, The percentage of transporter proteins encoded in the proteome in different species. d, The relative abundance of each type of metal transporter for each species. The legend in b is the same as for c and d. e, The percentage of Fe-, Mn- and Zn-binding proteins in the proteomes of Synechococcus, Emiliania huxleyi, Thalassiosira pseudonana and Thalassiosira oceanica versus half-saturation constants of these trace metals for the same phytoplankton species. The sizes of metalloproteomes in phytoplankton positively correlate with their metal requirements, indicated by the half-saturation constants of different trace metals for growth. f, The relative abundance of different transporter groups in phytoplankton (ncyanobacteria = 2, ngreen = 13, nred = 11). P values from two-tailed t-tests between the green and red lineages are shown (P < 0.05 highlighted in bold). g, Percentage of thiol-containing amino acids in Fe and Zn transport related proteins in different groups of phytoplankton (the numbers of sequences analysed in the group are labelled, with the P values from the one-way ANOVA test for each group shown higher up and P < 0.05 highlighted in bold). IRT, iron-regulated transporter; FTR, Fe transporter. Boxes in f and g mark the 25th and 75th percentiles of values for each group. The line in the middle of the box is the median. The whiskers show the maximum and minimum values of the group. The points show the outliers that exceed 1.5 × the interquartile range of the group. h, PCA of all different types of metal transporter for different species of phytoplankton. Different lineages of phytoplankton are clustered together.
Contrasting metal requirements of phytoplankton lineages
The biological requirement of elements varies in different species16. Although previous studies convincingly show that different lineages of phytoplankton systematically differed in trace metal content10,11,12, the discovery of various metal storage proteins and the plasticity in elemental stoichiometry under different environmental conditions17,18 questions whether intracellular trace metal quotas reflect true differences in usage. Here we embellish the difference in metal requirements with genomically grounded use of metals. We determined that the percentage of the metalloproteins encoded in phytoplankton genomes indeed reflects the physiological requirements for trace metals (Figs. 1, and 2, and Supplementary Discussion). This approach could harbour major uncertainties associated with expression levels of different metalloproteins, but a good correlation exists between the half-saturation coefficient Ku of various phytoplankton species as a measure of their metal requirements compared with the proportion of the genome dedicated metal-binding proteins (Fig. 2e). Ku is the concentration that can support a growth rate at one-half the maximum growth rate of phytoplankton. The percentages of Fe-, Mn- and Zn-binding proteins in phytoplankton positively correlate with their Ku of Fe, Mn and Zn, respectively (Fig. 2e). Given that the percentage of encoded metalloproteome seems to be a useful indicator of physiological metal requirements, we extended our analysis to a larger number of phytoplankton genomes to gain insight into contrasting trace metal requirements (Fig. 1). The significantly lower percentage of metalloproteins in the secondary endosymbiotic lineages indicates that they have lower metal requirements for trace metals Fe, Zn and Cu, compared with those of primary endosymbionts (P < 0.05; Fig. 1).
Despite the gap between the bioinformatically predicted function of genes and the phenotypic metal quota, our analysis here can illustrate the proportional resource investment in phytoplankton trace metal acquisition and use (Figs. 1–3 and Supplementary Fig. 2). This yields additional dimensions that cannot be obtained via metal quota analysis and provides further insights into the co-evolution of life and environment10,19.
There are no mitochondria-targeted Cu and Co proteins/transporters, and no plastid-targeted Mo and Mo (VI) transporters. Compared with the primary endosymbionts, the secondary endosymbionts have relatively smaller fractions of metals, including Zn, Fe, Cu and Co, that are plastid-targeted. ABC transporters were not included in the analysis for Zn, Fe, Cu and Mn, but were included for Co as they are considered one of the major groups of Co uptake transporters in phytoplankton. The plastid metalloproteins comprise a larger proportion of the Fe- (30–53%), Cu- (4–25%), Mn- (14–80%) and Co-binding (30–67%) proteins in the primary endosymbionts than those in the secondary endosymbionts (17–27% Fe, 0–8% Cu, 0–25% Mn and 0–50% Co), which could reflect a reduction of the plastid proteome during evolution subsequent to endosymbiosis52. Many specific transporters for trace metals, including those for Zn (30–70%), Fe (10–25%), Cu (17–100%) and Co (30–90%), are also plastid-targeted to facilitate metal transportation across the plastid membranes. A high proportion of Mo-containing proteins are mitochondria-targeted (43–80% in the primary endosymbionts and 30–75% in the secondary endosymbionts), but none of the Mo(VI) transport proteins are found to be specifically targeting the plastid or mitochondria.
Strategies for metal management in different phytoplankton
Phytoplankton use metal transport systems for metal acquisition and distribution. The amount of energy dedicated to the transport of trace metals may hint at chemistry changes in seawater. Principal component analysis (PCA) of metal-transporting protein families in aggregate (numbers of proteins in each metal-transporter family in phytoplankton species) confirmed that phytoplankton from the same phyla cluster together (Fig. 2h), with the first two components clearly separating cyanobacteria, red lineage and green lineage phytoplankton (Fig. 2h). The separation of phytoplankton into distinct groups from PCA analysis of metal transporters agrees with the separation via elemental stoichiometry11.
To compare the relative genetic investment of phytoplankton in transporting of different metals, we calculated the percentages of different metal transporters relative to the proteome sizes across the groups (Fig. 2). Cyanobacteria encode a significantly greater proportion (~2.8%) of metal transporters in the proteomes compared with eukaryotes (0.5–1%, P < 0.05; Fig. 2c). Adenosine triphosphate (ATP)-binding cassette transporters (ABC transporters), one of the largest and oldest gene families that are responsible for the ATP-powered translocation of many substrates across membranes20, are the most abundant metal-transporter families in all phytoplankton (Fig. 2f). Particularly, in cyanobacteria, they comprise more than 90% of all metal-transporter families—these transporters typically transport more than one type of metal, such as the Ni/Co uptake transporter family and the Mn/Zn/Fe chelate uptake transporter family20. The low diversity in transporter type (Supplementary Fig. 3) in cyanobacteria compared with eukaryotes reflects intense genome streamlining21. Moreover, some cyanobacteria lack phytochelatin synthase (Supplementary Table 2), known to mediate the synthesis of metal-binding phytochelatins22, a strategy for managing metal toxicity by binding and storing of excess metals23. Cyanobacteria have a low percentage of P-type ATPases, transporters that confer protection from extreme environmental stress conditions such as high concentrations of metals24,25. These features of cyanobacterial metal transport systems—a high investment (high percentage of transporter proteins in the proteome), streamlined and indiscriminate metal uptake and efflux system—indicates that they employ a strategy centred around nutrient acquisition and efflux, making cyanobacteria ubiquitously well adapted.
Among eukaryotic phytoplankton, the chlorophyll a+c-containing (red) lineages have a lower diversity in their transporter systems compared with the chlorophyll b-containing (green) lineages. Many of the red lineage phytoplankton lack specific Cu transporters (for example, the COPT group) and high-affinity Ni transporters, but encode a greater number of generalist transporters (for example, ABC transporters; Fig. 2f and Supplementary Fig. 3)20. The ABC transporters comprise a significantly greater proportion (P < 0.05) of metal transporters in the red lineages (60%) compared with green lineages (50%). In contrast to the red lineage phytoplankton, the green lineages have significantly higher percentages of P-type ATPase dedicated to metal efflux (Fig. 2f, P < 0.001)24,25. The green lineages have more complex metal transport systems, likely to eliminate an excess of specific metals from cells, and therefore may be better adapted for environments with metals at higher potentially toxic concentrations26,27 such as modern nutrient-rich coastal or upwelling environments28,29. Such an adaptation may contribute to the green algae providing the successful ancestors for the modern land plants, as living in the terrestrial realm requires the ability to select for essential metals from the metal-rich rock and soil milieu. In contrast, the red lineages have fewer metal transport systems comprising more general metal transporters, meaning that they are better adapted to an oligotrophic environment where the risk of uptake of excess metals is lower21.
The affinity of metal transporters, indicated by the proportions of sulfur- or thiol-containing amino acids that have high affinity for transition metals30, is greater in the secondary endosymbionts compared with the primary endosymbionts, in various metal-transporting proteins (Fig. 2g, Supplementary Fig. 4 and Supplementary Table 3), especially those related to Fe- and Zn-transporting processes, such as Zn transporters (for example, ZIPs) and ferric reductases, which reduce Fe(III) to Fe(II) for Fe transmembrane uptake. No systematic variations in the average thiol amino acid compositions were found in different algal lineages, suggesting that the differences in the thiol content found in the transporters are not resulting from genome-wide bias (Supplementary Fig. 4). The histidine contents are also found to be significantly higher in the high-affinity Ni transporting family, multicopper oxidases and the Fe(III) transporters (FTR family) in the secondary endosymbionts (Supplementary Table 3). This suggests that the secondary endosymbionts may have evolved a more selective metal transport system with improved discrimination against abundant metal competitors for the binding sites (for example, Mg and Ca) in seawater that are orders of magnitude higher than those essential trace elements such as Fe and Zn, supported by a higher sulfate availability in the marine environment31. Having a higher-affinity transporting system for essential trace metals Fe and Zn could be an advantageous feature for the secondary endosymbionts to live in the oligotrophic open ocean.
How differences in chemistry and strategies arise
We conducted a protein location analysis to test whether the evolutionary history of the plastid is the source of the differences in metal requirements and transport strategy of the cell. All orthogroups of metalloproteins and metal transporters, annotated to be plastid- or mitochondria-targeted, were extracted across different phytoplankton species for each trace element (Fig. 3). The variation in proportions of plastid-targeted metalloproteins and transporters of different phytoplankton groups partially explains the evolutionary inheritance of elemental requirements found in different eukaryotic marine phytoplankton10. Elements that were more bioavailable billions of years ago, such as Fe and Co, are integral to the metalloproteome of cyanobacteria. A higher proportion of these elements can be attributed to the plastids derived from cyanobacteria. In contrast, higher proportions of elements such as Mo, which were less bioavailable under anoxic ocean conditions, are found in proteins originating from eukaryotes (that is, these protein groups are absent from the cyanobacteria; Fig. 3). Although no significant difference in plastid core protein-coding genes was found at the superfamily level previously10,32, a reduction in proportions of plastid metalloprotein/transporter of various trace elements was found in the secondary-endosymbiont-bearing lineages, suggesting a metalloproteome reduction in the plastid via endosymbiosis and a potential increased contribution from the eukaryotic host in regulating their trace element requirement and transport ability, especially for elements such as Fe, Cu and Co.
Metal requirements may have coevolved with metal availability
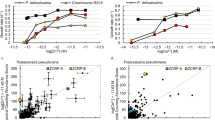
Contrasting metal use and uptake strategies for Fe and Zn, in particular, reveal how evolving ocean chemistry has shaped parallel changes in the metalloproteome and metal acquisition strategies of different lineages of phytoplankton (Fig. 1). These two metals comprise the largest proportion of the metalloproteome in various organisms and are involved in a more diverse range of biological pathways and activities than other essential trace metals33,34. Fe is an abundant metallocentre in photosynthesis and electron transfer (Supplementary Fig. 2), being found in almost all electron transfer complexes (photosystem II, photosystem I, cytochrome b6f complex and ferredoxins), and abundantly bound with plastid or mitochondria proteins in the eukaryotic phytoplankton (Fig. 3). Our analysis reveals an inverse relation between the proportions of specific Fe transporters in trace metal transporters with the percentage of Fe-binding proteins in phytoplankton proteomes (Fig. 4a and Supplementary Fig. 5a). The emergence of cyanobacteria in an anoxic ocean, with abundant soluble Fe(II) and continued selection to inhabit an enviroment with relatively greater Fe bioavailability35, is consistent with both the greater Fe-binding proportion of cyanobacteria proteomes and the lower proportion of high-affinity Fe-specific transporters, reflecting the absence of a selective pressure to develop a specific Fe uptake system. The later emergence of the red lineage secondary endosymbionts, such as diatoms, in an environment where the bioavailable Fe concentration was orders of magnitude lower, have been selected for streamlining Fe use in their proteomes. Meanwhile they developed more Fe transporters/pathways for Fe acquisition to fulfil this reduced Fe need in response to the scarcity of this key element.
a, Fe. b, Zn. ABC transporters were not included in this analysis. The shaded bands represent 95% confidence intervals for the fitted values based on a logarithmic regression of the data points.
A positive correlation exists between the proportion of specific Zn transporters and Zn-binding proteins in different phytoplankton proteomes (Fig. 4b and Supplementary Fig. 5b). Larger proportions of Zn in eukaryotic than prokaryotic proteomes are attributed to small Zn-binding domains such as Zn fingers and RING domains, involved in protein–DNA/RNA interactions and protein–protein interactions36. A positive linear relationship between the number of Zn-containing proteins and the total number of proteins (r2 = 0.91) of different phytoplankton (both cyanobacteria and eukaryotes; Supplementary Fig. 6) indicates an increase of Zn requirements with complexity of life, consistent with the previous finding that Zn in higher organisms is essential for many enzymatic activities and enabling a tight contol of gene expression33,36. The higher percentage of Zn in eukaryote proteomes compared with cyanobacteria and a higher number of Zn transporters to manage Zn homeostasis in their cells may reflect a possible increase of bioavailable Zn in the surface ocean based on thermodynamic considerations37. Such an increase in Zn is not well supported by current geological evidence1,38,39 (Supplementary Discussion), so either there are secondary imprints on the geological record or the increased use of Zn in eukaryotes may just reflect the uniqueness of its chemistry to life, leading to more extensive use and incorporation of Zn into more functions, enabling more complex life to evolve40.
Linking metal availability with phytoplankton shifts
Evolving ocean chemistry contributed selective pressure to promoting the proliferation of different phytoplankton groups through major transitions in Earth’s history8,41,42 (Fig. 5 and Supplementary Fig. 1), given their differences in metalloproteome and metal transport strategy. The surge of both major and minor nutrients in the surface ocean supplied by the Sturtian deglaciation41,42 may have promoted the rise of green algae (Chlorophyta, primary endosymbionts), well adapted to trace-metal-rich conditions. The contemporaneous increased atmospheric oxygenation43 may have contributed to reduced bioavailable Fe but increased bioavailable Zn in the surface ocean and aggravated the selection for the lower-Fe and higher-Zn requirements of the green algae compared with cyanobacteria (Fig. 1). Vertical distributions of metals may have been altered even if the Zn content of the whole ocean remained broadly uniform throughout time1,38,39. It was not until the Mesozoic that the secondary endosymbionts were able to radiate across oceanic shelves into the open ocean (Fig. 5 and Supplementary Fig. 1). They harbour a more metal-lean strategy, with low requirements of Fe, Cu and Zn in their proteomes, compared with the primary endosymbionts (Fig. 1). The more general transport strategies of the secondary endosymbionts compared with the selective transport systems of the primary endosymbionts also make the secondary endosymbionts better adapted to a trace-metal-poor, oligotrophic marine environment. The rise of the secondary endosymbionts to dominance at the Mesozoic therefore points to a distinct shift towards a paucity of trace metals in the open surface ocean ecological niche at this time, a target for validation by future geochemical records (Fig. 5 and Supplementary Fig. 7).
a, NO3 and PO4 concentrations are from the COPSE reloaded model53, atmospheric O2 level is from the GEOCARBSULFOR model54. b,c, Concentrations of Zn (b) and Cu (c) are from pyrite and shale records2,55. Each dot represents a pyrite (blue) or shale (grey) analysis and the lines represent the moving average of data for each 10 Myr interval. d, The Mesozoic marine revolution (shaded area) occurred during an extended interval of important evolutionary change in marine primary producers19, including radiations of photosynthetic dinoflagellates, coccolithophorids and diatoms. Ca/Mg ratios in seawater are model results from ref. 56.
The modern ocean allows a validation of this environmental selection by trace metals between niches of different phytoplankton groups, evidenced by their proteomes and geological succession. In the modern ocean, higher concentrations of trace metals, including Fe and Zn, are found in the coastal and upwelling zones, while the open ocean is metal scarce44. We extracted phytoplankton abundance data from the Ocean Biodiversity Information System and visualize them on the world ocean map (Fig. 6 and Supplementary Fig. 8). Although there are variations and uncertainties due to the differences in ecotypes within each group, different phytoplankton groups generally display contrasting ecologies consistent with their strategies for trace metal tolerance and acquisition. The secondary endosymbionts are dominant in the modern open ocean45 and, in most cases, the primary endosymbionts Chlorophyta and Rhodophyta are more abundant in the coastal regions32,46,47. The cell densities for cyanobacteria are abundant in both coastal and open-ocean regions, consistent with our prediction from metalloproteome and metal-transporter analysis. Although data regarding cyanobacterial transporters show that they employ a strategy centred around nutrient acquisition and efflux, allowing them to survive and be detectable in most modern open-ocean, nutrient-poor environments48, we also show that they have a greater Fe-binding proportion of the proteome, reflecting their high Fe requirement, supported by physiological experiments49,50 and metagenomics analysis, indicating their stress caused by Fe limitation in large areas in the open ocean35.
The points indicate the sampling location and colour represents the log scale of cell abundances (cells per litre). a, Cyanobacteria cell counts reach up to 107 cells l−1 in coastal areas (for example, St Lawrence estuary) and 105 cells l−1 in the open ocean (for example, North Atlantic Ocean). b, The primary endosymbiotic red lineage Rhodophyta is found mostly in the coastal region and the cell density can reach 106 cells l−1 (for example, Gulf of St Lawrence); its abundance is very low in the open ocean (for example, <1 cell l−1 in the Pacific Ocean). c, Abundances of the primary endosymbiont Chlorophyta. d, The cell densities of coccolithophores, the best-known haptophytes, can reach up to ~106 cells l−1 in the open ocean such as the mid-Atlantic, much higher than the primary endosymbionts Rhodophyta (b) and Chlorophyta (c), although we note that prasinophyceans, the pico-plankton with a size range of 0.2–2 µm in the Chlorophyta phylum, are also found in the open ocean with cell counts up to 103 cells l−1 (for example, North Pacific Ocean). Diatoms, another representative group of the secondary endosymbionts stramenopiles, are also found to be abundant in the open ocean (Supplementary Fig. 6).
Trace metal availability and thus their role in phytoplankton growth continues to evolve in the modern ocean. The contemporary ocean ecosystem is under threat from stressors associated with climate change, including warming, acidification, deoxygenation and pollution, which have implications for coastal eutrophication and the modulation of coastal versus open-ocean habitats. The change in trace metal geochemistry associated with the stressors may also substantially alter the phytoplankton community structure and thus carbon export activity in the future ocean51.
Methods
To ensure the robustness and validity of our conclusions, we have employed two different approaches of analysis of the trace metal complement of different phytoplankton in this study. The majority of the data are collected and analysed by a comparative genomic approach, using complete genome-inferred proteomes of different phytoplankton species. To augment the data, we conducted domain analysis on both the complete proteomes of the phytoplankton species and all sequences from the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP)57. In this analysis, we used the Structural Classification of Proteins (SCOP) database58 and MetalPDB database59 as our library and references for protein domain and metal domain annotations. The uncertainties associated with these two approaches are discussed in the Supplementary Information.
Proteome sources and raw data generation for the comparative genomics approach
In the comparative genomic analysis, to evaluate the proportional importance of each trace metal for the different lineages (Fig. 1), we calculated the percentage of metalloproteins binding with specific trace metals in the genome-predicted proteomes of 26 species of phytoplankton (Supplementary Table 1). Although our list of phytoplankton in this study is not exhaustive of the current phytoplankton proteomic database, the species included are from a diverse range of phyla from two superfamilies: the chlorophyll a+b-containing eukaryotes (green lineage) including Chlorophyceae, Trebouxiophyceae, Klebsomidiophyceae and Prasinophyceae; and the chlorophyll a+c-containing eukaryotes (red lineage) including Bangiophyceae, Florideophyceae, Cyanidiophyceae, Cryptophyta, Haptophyta and Stramenophiles. In the red lineage group, Cryptophyta, Haptophyta and Stramenopiles are secondary endosymbionts, while the others are primary endosymbionts. Although the focus of this study is the differences between the dominant eukaryotic phytoplankton groups, we have included two species of cyanobacteria, which are widely observed in the ocean and are major contributors to primary production35, for comparison.
Groups of orthologues shared among phytoplankton species were inferred using OrthoFinder60,61 with DIAMOND (double index alignment of next-generation sequencing data) for protein alignment62. In detail, we conducted all-versus-all DIAMOND blast throughout our list of phytoplankton sequences to measure pairwise sequence similarity between all sequences in the species. To reduce the impact of gene length on clustering accuracy, the method we used has employed a protocol that determines the gene length dependency of a given pairwise species comparison from an analysis of the bit scores from an all-versus-all Blast search60. Briefly, for each species pair, the all-versus-all Blast hits were divided into equal sized bins of increasing sequence length according to the product of the query and hit sequence length. The best 5% of hits were used to represent good hits for sequences of that length bin and a linear model in log–log space was used to fit a line to the scores by least square method. By transforming all the Blast bit scores from each species pair all-versus-all search using this model, the best hits between sequences in the species pair have equivalent scores that are independent of sequence length. Such score transformation enabled a normalization for both gene length and phylogenetic distance between species. These scores were used as the measure of sequence similarity on which all subsequent analysis and clustering were performed. After obtaining the orthogroups, the gene trees for all orthogroups are inferred and then from these gene trees the rooted species tree is identified. Then the method we employed provided both gene tree and species tree-level analysis of all gene duplication events. Based on all this phylogenetic information, we then can identify the complete set of orthologues between all species of phytoplankton that we investigated. Here, the orthogroup graph was clustered using Markov Cluster Algorithm (MCL) with its default inflation parameter of 1.5 (https://micans.org/mcl/). The species tree for the phytoplankton investigated in this study was also constructed using OrthoFinder based on the Species Tree from All Genes (STAG) algorithm, which infers the species tree using the most closely related genes within single-copy or multi-copy orthogroups. The output comparative genomics data and statistics were then subjected to further analysis for investigating metal transporters and metalloproteins. We have searched and identified the metal-binding proteins according to their functions, that is, metal transporters or metalloproteins that have other functions. We aim to understand the usage of trace metals by phytoplankton as well as their strategies for transporting different metals. These two processes together can determine the trace metal requirements by phytoplankton as reflected in their responses to trace metal limitation and toxicity. For each metalloprotein, we take into consideration of all different metals that can work as co-factors. For instance, Cu/Zn superoxide dismutase would be considered both a Cu metalloprotein and a Zn metalloprotein, to reflect its potential use of these two different trace metals.
We use annotations from previous studies on model organisms as our library and references (listed in Supplementary Table 1). Most sequences used in this analysis are reference proteomes from the Universal Protein Resource (UniProt). They were rigorously assembled and annotated by experts in the field and were selected as landmarks in proteome space. Moreover, because we have conducted comparative genomics analysis here, the annotations of the orthogroups from all phytoplankton species included in this analysis are compared and cross-checked against those from all other species; that is, even if we have proteins that derive from relatively worse-annotated species, they are further checked and compared with those better-annotated species to enable a robust analysis in our study.
Protein (metal) domain analysis
In the domain level analysis, in addition to all the phytoplankton species from UniProt shown in Supplementary Fig. 1, we included whole proteome sequences of four phytoplankton species that have not been analysed in the comparative genomic approach: the stramenopile Phaeodactylum tricornutum and dinoflagellates Polarella glacialis, Symbiodinium microadriaticum and Symbiodinium pilosum. These sequences are also all from UniProt. We have also analysed all available sequences from the MMETSP. To do that, we applied a DIAMOND Blast search of all the sequences against the SCOP database to identify all the domains for each species, using an Expect value (E) of 10−4 as the cut-off. Then we searched the Protein Data Bank ID for each domain against the MetalPDB database to identify all the metal-containing domains. The relative abundances of trace metals for each species were then calculated by using the total number of metal domains normalized to total number of protein domains identified. We then conducted statistical analysis for different groups of phytoplankton to understand their relative requirement of trace metals. As the quality of the sequences from the MMETSP varies from strain to strain, we included only those (a total of 608 sequences) that had more than 200 protein domains identified for further metal domain and statistical analysis.
Phytoplankton half-saturation coefficient data
Ku (the half-saturation coefficient for growth) values of different phytoplankton can be used as a measure of their metal requirements. Ku is [S] when μ = μmax/2 as described by the response of phytoplankton physiology (growth) to external trace metal concentrations using the steady-state equation for substrate limitation (Monod equation, equation (1):
wherein [S] is the concentration of the limiting nutrient; μ is the specific growth rate and μmax is the maximum specific growth rate. The raw data for calculating the half-saturation constants are from ref. 63. Only data from the species that overlapped with our metalloproteome analysis are included in Fig. 2, including Synechococcus, Emiliania huxleyi, Thalassiosira pseudonana and Thalassiosira oceanica. We used data from a single study in which all conditions were consistent to avoid potential differences in Ku values caused by variations in experimental conditions.
Data statistical analysis
Metal-transporting families and metalloproteins analysis
For each metal of interest, we searched across the data generated by OrthoFinder and extracted all data annotated as metal-containing or transporting families in the proteome. We constructed matrices with each column representing a distinct species and each row representing either the abundances of metal-related proteins or the detailed proteins accessions in the group. To determine whether the encoded metalloprotein abundances were different between phyla and subgroups of phytoplankton, we performed a series of t-tests and analysis of variance (ANOVA) on each dataset. Patterns in metal-transporter systems were also examined using PCA. Multivariate clustering on different matrices were performed using Past (https://uhm.uio.no/english/research/resources/past/).
Amino acid composition of transporter families
We extracted protein sequences for each transporter family from all phytoplankton included in this study and determined their amino acid compositions by counting the elements forming each amino acid in the transporter protein sequences. For each type of transporter, the data were separated into phytoplankton subgroups, including the cyanobacteria, the Rhodophyta, the Chlorophyta and the secondary endosymbionts. Single-factor one-way ANOVA analysis was employed to determine the statistical differences among groups.
Distribution of phytoplankton
The abundance data of different phytoplankton in Fig. 6 were taken from the Ocean Biodiversity Information System (OBIS), which provides the world’s largest scientific knowledge base on the diversity, distribution and abundance of all marine organisms. The data for diatoms (Supplementary Fig. 6) are from ref. 64. We extracted data according to their phylum names and included only data with individual counts available in the analysis. Individual counts from each data point were then plotted on a 3 × 3° global grid base map using the Generic Mapping Tools65.
Data availability
All original data from this study can be obtained from publicly available databases. Completed proteome sequences used in this study are from UniProt (https://www.uniprot.org/) and the National Center for Biotechnology Information (https://ncbi.nlm.nih.gov). Sequences from the MMETSP can be accessed from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA248394). Structural domains for different protein families can be obtained via the SCOP database (http://scop.mrc-lmb.cam.ac.uk/). Details for metalloproteins can be found at MetalPDB (https://metalpdb.cerm.unifi.it/). Geological records of trace elements can be obtained from the Sedimentary Geochemistry and Paleoenvironments Project database (https://sgp.stanford.edu/). Cell abundance data for different groups of phytoplankton are from OBIS (https://obis.org/). All analysed data in this study can be download via https://dataspace.hkust.edu.hk/dataverse/metalloproteins. Source data are provided with this paper.
References
Robbins, L. J. et al. Trace elements at the intersection of marine biological and geochemical evolution. Earth-Sci. Rev. 163, 323–345 (2016).
Large, R. R. et al. Cycles of nutrient trace elements in the Phanerozoic ocean. Gondwana Res. 28, 1282–1293 (2015).
Long, J. A. et al. Severe selenium depletion in the Phanerozoic oceans as a factor in three global mass extinction events. Gondwana Res. 36, 209–218 (2016).
Lu, W. et al. Late inception of a resiliently oxygenated upper ocean. Science 361, 174–177 (2018).
Strassert, J. F. H., Irisarri, I., Williams, T. A. & Burki, F. A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids. Nat. Commun. 12, 1879 (2021).
Rickaby, R. E. M. & Eason Hubbard, M. R. Upper ocean oxygenation, evolution of RuBisCO and the Phanerozoic succession of phytoplankton. Free Radic. Biol. Med. 140, 295–304 (2019).
Kodner, R. B., Pearson, A., Summons, R. E. & Knoll, A. H. Sterols in red and green algae: quantification, phylogeny, and relevance for the interpretation of geologic steranes. Geobiology 6, 411–420 (2008).
Brocks, J. J. The transition from a cyanobacterial to algal world and the emergence of animals. Emerg. Top. Life Sci. 2, 181–190 (2018).
Schwark, L. & Empt, P. Sterane biomarkers as indicators of Palaeozoic algal evolution and extinction events. Palaeogeogr. Palaeoclimatol. Palaeoecol. 240, 225–236 (2006).
Quigg, A. et al. The evolutionary inheritance of elemental stoichiometry in marine phytoplankton. Nature 425, 291–294 (2003).
Quigg, A., Irwin, A. J. & Finkel, Z. V. Evolutionary inheritance of elemental stoichiometry in phytoplankton. Proc. R. Soc. B 278, 526–534 (2011).
Finkel, Z. V., Quigg, A. S., Chiampi, R. K., Schofield, O. E. & Falkowski, P. G. Phylogenetic diversity in cadmium: phosphorus ratio regulation by marine phytoplankton. Limnol. Oceanogr. 52, 1131–1138 (2007).
Waldron, K. J. & Robinson, N. J. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7, 25–35 (2009).
Hanikenne, M. A comparative inventory of metal transporters in the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschizon merolae. Plant Physiol. 137, 428–446 (2005).
Boden, J. S., Konhauser, K. O., Robbins, L. J. & Sánchez-Baracaldo, P. Timing the evolution of antioxidant enzymes in cyanobacteria. Nat. Commun. 12, 4742 (2021).
Frausto da Silva, J. J. R. & Williams, R. J. P. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life (Oxford Univ. Press, 2001).
Maret, W. & Wedd, A. Binding, Transport and Storage of Metal Ions in Biological Cells (Royal Society of Chemistry, 2014).
Galbraith, E. D. & Martiny, A. C. A simple nutrient-dependence mechanism for predicting the stoichiometry of marine ecosystems. Proc. Natl Acad. Sci. USA 112, 8199–8204 (2015).
Falkowski, P. G. et al. The evolution of modern eukaryotic phytoplankton. Science 305, 354–360 (2004).
Zheng, W. H. et al. Evolutionary relationships of ATP-binding cassette (ABC) uptake porters. BMC Microbiol. 13, 98 (2013).
Hogle, S. L., Cameron Thrash, J., Dupont, C. L. & Barbeau, K. A. Trace metal acquisition by marine heterotrophic bacterioplankton with contrasting trophic strategies. Appl. Environ. Microbiol. 82, 1613–1624 (2016).
Blum, R., Meyer, K. C., Wünschmann, J., Lendzian, K. J. & Grill, E. Cytosolic action of phytochelatin synthase. Plant Physiol. 153, 159–169 (2010).
Cobbett, C. S. Heavy metal detoxification in plants: phytochelatin biosynthesis and function. IUBMB Life 51, 183–188 (2001).
Chan, H. et al. The P-type ATPase superfamily. J. Mol. Microbiol. Biotechnol. 19, 5–104 (2010).
Hanikenne, M. & Baurain, D. Origin and evolution of metal P-type ATPases in Plantae (Archaeplastida). Front. Plant Sci. 4, 544 (2014).
Wilson, W., Zhang, Q. & Rickaby, R. E. M. Susceptibility of algae to Cr toxicity reveals contrasting metal management strategies. Limnol. Oceanogr. 64, 2271–2282 (2019).
Zhang, Q. & Rickaby, R. E. M. Interactions of thallium with marine phytoplankton. Geochim. Cosmochim. Acta 276, 1–13 (2020).
Berger, W. H., Smetacek, V. & Wefer, G. in Productivity of the Ocean: Present and Past (eds Berger, W. H. et al.) 1–34 (John Wiley, 1989).
Butterfield, N. J. Plankton ecology and the proterozoic-phanerozoic transition. Paleobiology 23, 247–262 (1997).
Bozzi, A. T. et al. Conserved methionine dictates substrate preference in Nramp-family divalent metal transporters. Proc. Natl Acad. Sci. USA 113, 10310–10315 (2016).
Ratti, S., Knoll, A. H. & Giordano, M. Did sulfate availability facilitate the evolutionary expansion of chlorophyll a+c phytoplankton in the oceans? Geobiology 9, 301–312 (2011).
Grzebyk, D., Schofield, O., Vetriani, C. & Falkowski, P. G. The Mesozoic radiation of eukaryotic algae: the portable plastid hypothesis. J. Phycol. 39, 259–267 (2003).
Dupont, C. L., Yang, S., Palenik, B. & Bourne, P. E. Modern proteomes contain putative imprints of ancient shifts in trace metal geochemistry. Proc. Natl Acad. Sci. USA 103, 17822–17827 (2006).
Sevcenco, A. M., Hagen, W. R. & Hagedoorn, P. L. Microbial metalloproteomes explored using MIRAGE. Chem. Biodivers. 9, 1967–1980 (2012).
Ustick, L. J. et al. Metagenomic analysis reveals global-scale patterns of ocean nutrient limitation. Science 372, 287–291 (2021).
Andreini, C., Banci, L., Bertini, I. & Rosato, A. Zinc through the three domains of life. J. Proteome Res. 5, 3173–3178 (2006).
Saito, M. A., Sigman, D. M. & Morel, F. M. M. The bioinorganic chemistry of the ancient ocean: the co-evolution of cyanobacterial metal requirements and biogeochemical cycles at the Archean–Proterozoic boundary? Inorg. Chim. Acta 356, 308–318 (2003).
Scott, C. et al. Bioavailability of zinc in marine systems through time. Nat. Geosci. 6, 125–128 (2013).
Robbins, L. J. et al. Authigenic iron oxide proxies for marine zinc over geological time and implications for eukaryotic metallome evolution. Geobiology 11, 295–306 (2013).
Williams, R. J. P. & Rickaby, R. E. M. Evolution’s Destiny: Co-evolving Chemistry of the Environment and Life (Royal Society of Chemistry, 2012).
Brocks, J. J. et al. The rise of algae in Cryogenian oceans and the emergence of animals. Nature 548, 578–581 (2017).
Gueneli, N. et al. 1.1-billion-year-old porphyrins establish a marine ecosystem dominated by bacterial primary producers. Proc. Natl Acad. Sci. USA 115, E6978–E6986 (2018).
Lyons, T. W., Reinhard, C. T. & Planavsky, N. J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315 (2014).
Moore, C. M. et al. Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710 (2013).
de Vargas, C. et al. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015).
Falkowski, P. G., Schofield, O., Katz, M. E., Van de Schootbrugge, B. & Knoll, A. H. in Coccolithophores (eds Thierstein, H. R. & Young, J. R.) 429–453 (Springer, 2004).
Katz, M. E., Finkel, Z. V., Grzebyk, D., Knoll, A. H. & Falkowski, P. G. Evolutionary trajectories and biogeochemical impacts of marine eukaryotic phytoplankton. Annu. Rev. Ecol. Evol. Syst. 35, 523–556 (2004).
Rusch, D. B., Martiny, A. C., Dupont, C. L., Halpern, A. L. & Venter, J. C. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc. Natl Acad. Sci. USA 107, 16184–16189 (2010).
Berman-Frank, I., Quigg, A., Finkel, Z. V., Irwin, A. J. & Haramaty, L. Nitrogen-fixation strategies and Fe requirements in cyanobacteria. Limnol. Oceanogr. 52, 2260–2269 (2007).
Brand, L. E. Minimum iron requirements of marine phytoplankton and the implications for the biogeochemical control of new production. Limnol. Oceanogr. 36, 1756–1771 (1991).
Teng, Y. C., Primeau, F. W., Moore, J. K., Lomas, M. W. & Martiny, A. C. Global-scale variations of the ratios of carbon to phosphorus in exported marine organic matter. Nat. Geosci. 7, 895–898 (2014).
Keeling, P. J. The endosymbiotic origin, diversification and fate of plastids. Phil. Trans. R. Soc. B 365, 729–748 (2010).
Lenton, T. M., Daines, S. J. & Mills, B. J. W. COPSE reloaded: an improved model of biogeochemical cycling over Phanerozoic time. Earth-Sci. Rev. 178, 1–28 (2018).
Krause, A. J. et al. Stepwise oxygenation of the Paleozoic atmosphere. Nat. Commun. 9, 4081 (2018).
Farrell, Ú. C. et al. The Sedimentary Geochemistry and Paleoenvironments Project. Geobiology 19, 545–556 (2021).
Berner, R. A. A model for calcium, magnesium and sulfate in seawater over Phanerozoic time. Am. J. Sci. 304, 438–453 (2004).
Keeling, P. J. et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12, e1001889 (2014).
Andreeva, A., Kulesha, E., Gough, J. & Murzin, A. G. The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 48, D376–D382 (2020).
Andreini, C., Cavallaro, G., Lorenzini, S. & Rosato, A. MetalPDB: a database of metal sites in biological macromolecular structures. Nucleic Acids Res. 41, D312–D319 (2018).
Emms, D. M. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2014).
Brand, L., Sunda, W. G. & Guillard, R. R. L. Limitation of marine phytoplankton reproductive rates by zinc, manganese, and iron. Limnol. Oceanogr. 28, 1182–1198 (1983).
Leblanc, K. et al. A global diatom database - abundance, biovolume and biomass in the world ocean. Earth Syst. Sci. Data 4, 149–165 (2012).
Wessel, P. et al. The generic mapping tools version 6. Geochem. Geophys. Geosyst. 20, 5556–5564 (2019).
Acknowledgements
We thank M. Tian (University of Bern), F. Wang (University of Oxford) and Z. Huang (Sun Yat-Sen University) for assistance with statistic analysis, and D. Porceilli (University of Oxford) for advice on geochemical data interpretation. We thank the following for funding: ERC Consolidator Grant APPLES (ERC-2015-COG-681746 to R.E.M.R.); Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) SMSEGL20SC01 to Q.Z.; Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) SML2021SP204 to Q.Z.; UK Natural Environmental Research Council (NERC) to R.S.; Guangdong Province Introduced Innovative R&D Team of Geological Processes and Natural Disasters around the South China Sea (2016ZT06N331 to Y.Z.); Deep Earth Exploration and Resource Environment (2017ZT07Z066 to Y.Z.); Portuguese Foundation for Science and Technology (CEECIND/00229/2018 to B.N.); and the Royal Society for a Wolfson Research Merit Award to R.E.M.R.
Author information
Authors and Affiliations
Contributions
Q.Z and R.E.M.R. designed the study; Q.Z conducted the study and wrote the initial draft; E.M.B, Y.Z. and B.N contributed to methodology developments; R.S contributed to discussion; all authors contributed to data interpretation and writing/editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Geoscience thanks Leslie Robbins and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: James Super, in collaboration with the Nature Geoscience team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Discussion, Supplementary Tables 1–3 and Supplementary Figures 1–8.
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 6
Statistical Source Data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, Q., Bendif, E.M., Zhou, Y. et al. Declining metal availability in the Mesozoic seawater reflected in phytoplankton succession. Nat. Geosci. 15, 932–941 (2022). https://doi.org/10.1038/s41561-022-01053-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41561-022-01053-7
This article is cited by
-
Phytoplankton in the middle
Nature Geoscience (2022)